3130
A Deep Learning Approach to Synthesize FLAIR Image from T1WI and T2WI1Department of Radiological Sciences, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
Synopsis
We synthewized FLAIR images of the brain from T1WI and T2WI by using autoencoder, which is one of the state of the art deep-learning technology. Autoencoder compresses the input information and reproduces the information therefrom. We used T1WI and T2WI as an input and synthewized FLAIR image with high accuracy. This method could be applicable to other body part other than the brain and might synthewize of other MR imaging sequences. This technology seems to be useful to improve clinical diagnosis and computer-aided diagnosis.
INTRODUCTION
FLAIR is an essential imaging sequence for the interpretation of the Neurological diseases of the brain. Sick patients may not be able to tolerate for a long scan time. To simplify the scan time and synthesize FLAIR images from basic T1WI, T2WI using state-of-the-art deep learning technology will help patients in daily clinical practice.METHODS
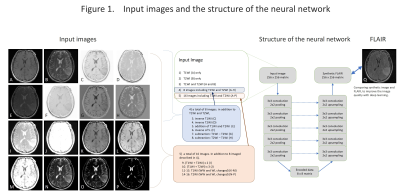
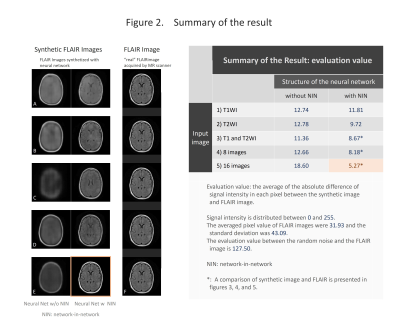
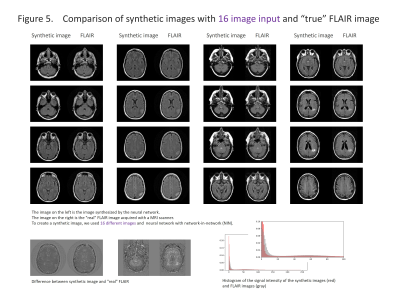
We selected the cases with 2D-T1WI, 2D-T2WI and 2D-FLAIR from the TCGA-GBM database. Fifty-two cases were chosen and 3414 images were used for the analysis. The images were reconstructed to 256×256 pixels and stored as a gray-scale png image (8-bit data, that means signal intensity ranging from 0 to 255) with default setting of WW and WL. T1WI and T2WI were used as an input and we used a data augmentation described below: 1). T1WI only, 2). T2WI only, 3). T1WI and T2WI, 4) In addition to T1WI and T2WI, addition/subtraction of images, total 8 different images, 5). In addition to 4), WW/WL adjusted images, total of 16 types of inputs (figure 1). Random noise was added to the input data. We used two types of convolutional autoencoder, both has repeated 5 times of 3×3 convolution and 2×2 pooling, but one has network in network (NIN) [1] in the same network and the other has not. Sixty four different feature maps were acquired in each convolution. The output layer was initialized by the method reported by Glorot and Bengio [2] and sigmoid was used as the activation function. Other layers were initialized by the method reported by He et al. [3] and rectified linear unit was used as activation function. We performed batch normalization on all layers [4]. We used cross entropy error for loss function and optimized with Adam [5] and performed 200 epochs of training. We programed with python 3.5 installed on Ubuntu 14.04 operating system and used Keras 1.2.2, TensorFlow 1.0, CUDA toolkit 8.0 and CuDNN v5.1 as a framework for deep learning. For evaluation, we compared the absolute difference of signal intensity between the synthetic image and the FLAIR image by pixel-by-pixel, and the average of the difference was used as the evaluation.RESULTS
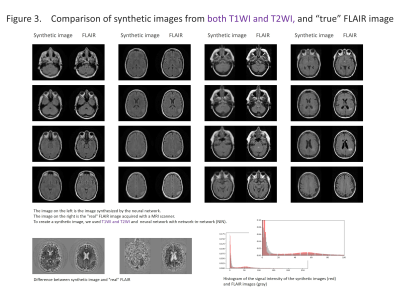
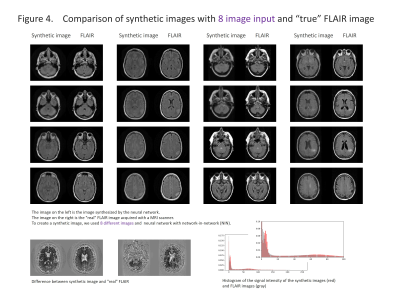
The results are shown in the figure 2-5 and summarized in the figure 2. NIN was crucial to obtain a high definition image (figure 2). The neural network with NIN with 16 different image inputs showed the best results (figure 5) and images synthetized with more different types of image were more similar to FLAIR image.
DISCUSSION
In this study, we found that FLAIR images could be created with high accuracy from T1WI and T2WI. Each institution acquires different imaging sequences due to the limitation of the scan time, and sometimes images are suboptimal for the image interpretation because of the poor quality due to motion artifact. This synthetic image may help shorten the scan time, and this method is further applicable for producing other sequences, like, T1WI, T2WI, fsT1WI, fsT2WI and STIR images. With the recent development of deep learning, research on computer-aided diagnosis (CAD) is rapidly progressing. In deep learning, a large amount of data is required to make high-precision CAD, but there are cases in which data cannot be used due to lack of some sequences. By using this technique, we can produce synthetic MR images, and the result will help further development of CAD research, which would be helpful to diagnostic radiologists. In this study, T1WI and T2WI are not enough for creating high precision synthetic FLAIR image. We found adding image modification factor can improve image quality; These include inversion, addition and subtraction of images, and adjustment of WW/WL, although this process didn’t increase the information of images. In addition, more complicated network rather than simple autoencoder shows better quality. In summary, quality of synthetic FLAIR image might be improved by using different image modification and a more complicated neural network.CONCLUSION
Our result demonstrated high definition synthetic FLAIR images can be created from T1WI and T2WI. This method has clinical implication to improve diagnosis and CAD optimization.Acknowledgements
We would like to acknowledge the individuals and institutions that have provided TCGA-GBM database.References
[1] Lin M, Chen Q, Yan S. Network in network. arXiv preprint arXiv:13124400 2013.
[2] Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, 2010; p. 249-56.
[3] He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. Proceedings of the IEEE international conference on computer vision, 2015; p. 1026-34.
[4] Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. International Conference on Machine Learning, 2015; p. 448-56.
[5] Kingma D, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980 w2014.
Figures