3123
Atlas-based brain extraction for bias field-affected structural imaging demonstrated using MPRAGE in marmoset1Signal and Image Processing Group, Laboratory of Metabolic Imaging, Singapore Bioimaging Consortium, Singapore, Singapore, 2MR Methods Development Group, Laboratory of Metabolic Imaging, Singapore Bioimaging Consortium, Singapore, Singapore, 3Functional Metabolism Group, Laboratory of Metabolic Imaging, Singapore Bioimaging Consortium, Singapore, Singapore, 4Department of Diagnostic Radiology, Singapore General Hospital, Singapore, Singapore, 5Laboratory of Microbial Immunity, Singapore Immunology Network, Singapore, Singapore
Synopsis
Changes in volume and shape of structures under an intervention or disease state can be measured in many subjects by automatic segmentation of structural imaging. However, unwanted intensity variation (bias field artefact), such as observed using a surface coil, can compromise such segmentation. We investigated how this artefact could be resolved using post-processing, to yield accurate brain extraction from MPRAGE acquisition in a marmoset. This atlas-based method (ASM) significantly improved brain extraction, correcting multiple inaccuracies of the initial FSL-based method (FSM) such as exclusion of the olfactory bulb and inferior cerebellar structures, and is robust to bias field artefact.
Introduction
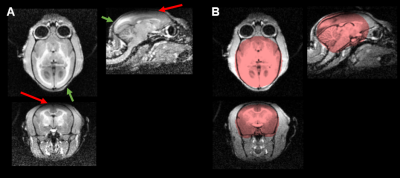
Structural imaging is used to detect changes in volume and shape of structures under an intervention or disease state. Manual segmentation is laborious, subjective to operator errors and impractical for large cohort studies. Automatic segmentation overcomes these limitations and is preferred for longitudinal studies. Magnetic field inhomogeneity, coil insensitivities and bias field affect the quality of imaging as well automatic segmentation of the structures. Imaging sequences such as T1-weighted MPRAGE1 are highly sensitive to bias field artefacts and field inhomogeneities, which can result in poor contrast, reducing the accuracy of brain extraction techniques (Figure 1). In this study we investigated how the surface coil bias field artefact could be resolved using post-processing to yield accurate brain extraction from MPRAGE acquisition in a group of marmosets.Methods
3D T1-weighted MPRAGE1 structural image of marmoset brain was acquired in 10 subjects (TI/TR/TE=900ms/2250ms/4.64ms ; NEX=3; FA=9°; Matrix=128x128x96; Res=0.4mmx0.4mmx0.4mm) on a Siemens Skyra 3T. The body volume coil was used as transmit coil and a 4cm single-channel surface coil as receiver coil. The receiver coil was positioned over the vertex of the marmoset head and secured using a custom rig. Marmoset was anaesthetized during data acquisition. The study was approved by Internal Ethics committee. Segmentation results were compared between the standard FSL3-based method (FSM) (Brain Extraction Tool2, f/g=0.8/0, robust mode) and the atlas-based method (ASM) outlined as follows:
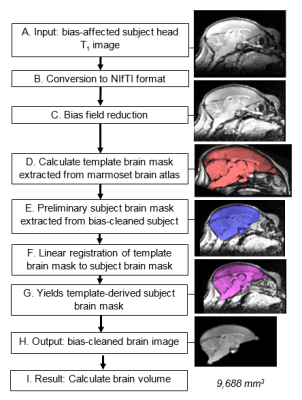
Proposed framework: The magnitude-reconstructed 3D volume was processed using the pipeline (Figure 2). First bias field reduction was performed using the mrbias tool from the Computational Morphometry Toolkit (CMTK)4. Brain extraction was then performed using the atlasBREX5 shell script, which uses FSL3 to register an atlas with a defined subject brain. Atlas developed by Hikishima et al.6 was used as a reference to estimate a preliminary subject brain-mask (Step E). Registration to the template brain-mask, yielded a template-derived subject brain-mask (Step G) from which brain volume could be measured (Step I).
Results
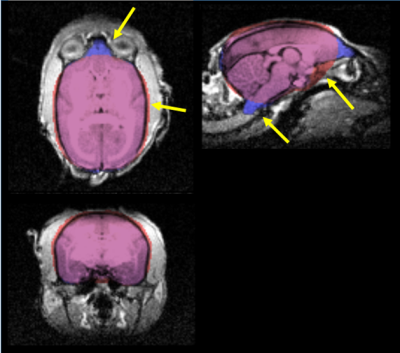
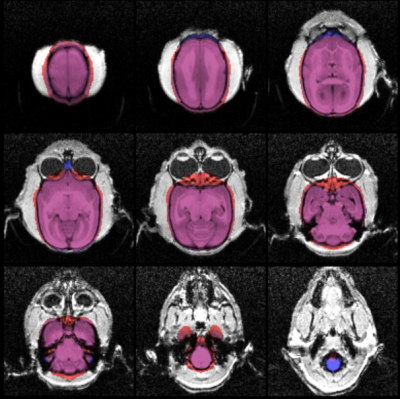
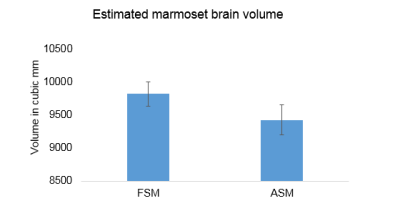
A 3D comparison of FSM (red) and ASM (blue) brain extraction is shown in Figure 3. Slice-by-slice comparisons in example axial slices are shown in Figure 4. All the brain extraction results were verified manually by neuroradiologist to check accuracy. Over 10 subjects, FSM yielded a significantly (paired t-test, p<0.05) overestimated (~4%) average brain volume of 9823±184mm3 in comparison to 9429±230mm3 from ASM (Figure 5). Structures that were missing in the FSM such as the olfactory bulb and brain stem were included in the ASM. Extraneous material, particularly in the inferior, lateral and superior sections, was removed by the ASM, which also correctly omitted the eyes.Discussion
The ASM significantly improved brain extraction, which was verified by visual inspection. It corrected many anatomical inaccuracies of the FSM based brain extraction by including intricate boundaries of the brain, the olfactory bulb and the inferior structures of the cerebellum and brain stem. While FSM is commonly used and reliable in conventional human studies, it had lesser accuracy on marmoset brain extraction. Bias field inhomogeneity had a stronger influence on brain extraction accuracy in FSM, which consistently manifested as underestimation of brainstem and olfactory bulb volumes, inclusion of extraneous anterior and inferior tissues, and skull tissue superior to the brain. ASM avoids dependence on intensity and bias field artefacts. By using the brain shape from the atlas, and registering this to the subject, the correct shape of the brain is preserved.Conclusion
A pipeline is demonstrated which successfully yields accurate brain volume measurements for small animals, despite the presence of severe bias field artefact from the surface ring coil. This extends existing analysis tools such as FSL to carry out atlas-based segmentation which is robust to presence of artefact in such studies.Acknowledgements
Agency for Science, Technology and Research (A*STAR) and Singapore General Hospital.References
1. Mugler JP, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magnetic Resonance in Medicine. 1990;15(1):152–157. 2. Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. 3. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004 [accessed 2017 Oct 19];23:S208–S219. http://www.sciencedirect.com/science/article/pii/S1053811904003933?via%3Dihub 4. Rohlfing T, Maurer CR. Nonrigid image registration in shared-memory multiprocessor environments with application to brains, breasts, and bees. IEEE transactions on information technology in biomedicine. 2003;7(1):16–25. 5. Lohmeier J, https://github.com/jlohmeier/atlasBREX. atlasBREX. 2017. 6. Hikishima K, Quallo MM, Komaki Y, Yamada M, Kawai K, Momoshima S, Okano HJ, Sasaki E, Tamaoki N, Lemon RN, et al. Population-averaged standard template brain atlas for the common marmoset ( Callithrix jacchus ). NeuroImage. 2011;54(4):2741–2749.Figures