3120
Pre-surgical high-resolution microstructural imaging in mesial temporal lobe epilepsy1Laboratory of Neuro Imaging, USC Stevens Neuroimaging and Informatics Institute, Keck school of medicine of USC, University of Southern California, Los Angeles, CA, United States, 2Department of Radiology, University of Southern California, Los Angeles, CA, United States
Synopsis
The gold standard for the treatment of medically refractory temporal lobe epilepsy continues to be surgical resection. This technique is not significantly different from when it was first popularized by Wilder Penfield in 1952. Significant advances in treatment are limited by our understanding of the structural abnormalities within the hippocampus prior to resection. In addition, pre-surgical planning for minimized resection demand accurate localization of hippocampal sclerosis (HS), which is limited by the achievable neuroimaging resolution. With advances in structural and diffusion MRI, microstructural imaging of brain tissue in high resolution is made possible, which can aid pre-surgical planning.
Purpose
In this work, we show the great potential of high-resolution diffusion microstructural imaging in pre-surgical planning of temporal lobectomy, for an accurate localization of hippocampal sclerosis (HS).Method
In this project, we used a Siemens 3T Prisma scanner to obtain pre-surgical structural and diffusion MRIs of a patient with mesial temporal lobe epilepsy (23-year old male). We then used a Bruker 16.4T scanner and imaged a 15 mm section of the dissected hippocampus. in vivo diffusion MRI was acquired using RESOLVE1 sequence, with in-plane resolution of 0.6 mm and 2 mm slice thickness. Two shells (800 and 2000 s/mm2) with total of 45 diffusion-encoding gradient directions were acquired. T2w image was acquired using BLADE2, with in-plane resolution of 0.4 mm and slice thickness of 2 mm. We also acquired MP2RAGE3 image with isotropic voxels of 1 mm. Total scan time was 60 minutes (44 and 9 minutes for RESOLVE and BLADE). For ex vivo MRI, multi-shell diffusion MRI and structural images were acquired with isotropic voxel sizes of 100 m and 50 m, respectively (total scan time of ~60 hours).Data Analysis
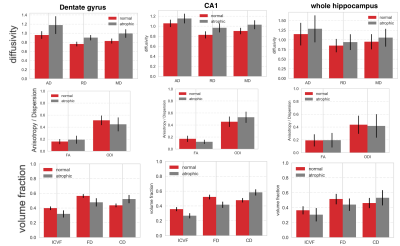
An adaptive optimized non-local mean filtering was applied on all the raw images to decrease the noise while avoid over-smoothing the data. Structural images were qualitatively compared with the MP2RAGE. Meso- and Micro-structural properties of hippocampus were compared between the atrophic and normal hippocampi (across entire hippocampus, CA1 and dentate gyrus). Models of diffusion tensor imaging (DTI4) and neurite orientation distribution and density imaging (NODDI)5 were fitted to voxels of hippocampus. In addition, NODDI measures were used to derive fiber and cellular densities using ABTIN6 model.Results
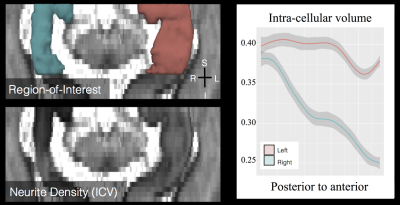
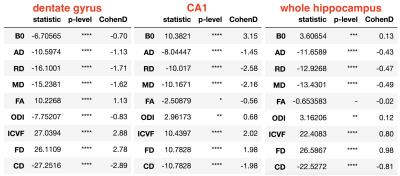
While an overall reduction in hippocampal size can be observed using MP2RAGE (at 1 mm), hippocampal subfields were unidentifiable (Figure 1). BLADE could resolve the subfields and showed that CA1 was the most atrophic. This was confirmed with ex vivo images. While BLADE offered a confident selection of the atrophic side, it could not provide microstructural information about the subfields, and therefore has limited localization application. Measures derived from the multi-compartment microstructural imaging (i.e. ICVF) could localize the HS with great accuracy, showing a high ICVF reduction in the anterior segment of the hippocampus (Figure 2). It also had the highest mean differences between the atrophic and normal hippocampi (Figure 3 and Table 1).Conclusion and future works
A more accurate localization of the HS can be achieved with high in-plane resolution imaging compare to conventional approaches. Tissue density measures from multi-compartment modeling (e.g. ICVF and FD) were more sensitive to HS compare to DTI and T2 contrast. These maps could enable better surgical planning and may be used as markers to evaluate alternative treatments. Our future focus would be on exploring ex vivo data and histological validation.Acknowledgements
No acknowledgement found.References
1. Porter, D. A., & Heidemann, R. M. (2009). High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magn Reson Med, 62(2), 468–475.
2. Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med 1999; 42:963-969.
3. Marques, J. P., Kober, T., Krueger, G., van der Zwaag, W., Van de Moortele, P. F., & Gruetter, R. (2010). MP2RAGE, a self bias-field corrected sequence for improved segmentation and T 1-mapping at high field. Neuroimage, 49(2), 1271-1281.
4. Basser, P. J., Mattiello, J., & LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophysical journal, 66(1), 259-267.
5. Zhang, H., Schneider, T., Wheeler-Kingshott, C. A., & Alexander, D. C. (2012). NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage, 61(4), 1000-1016.
6. Sepehrband, F., Clark, K. A., Ullmann, J. F., Kurniawan, N. D., Leanage, G., Reutens, D. C., & Yang, Z. (2015). Brain tissue compartment density estimated using diffusion‐weighted MRI yields tissue parameters consistent with histology. Human brain mapping, 36(9), 3687-3702.
Figures