3116
Towards Non-Invasive Characterization of Intravoxel Tumor Heterogeneity: Correlation between Non-Gaussian Diffusion MRI and Histology Using Machine Learning1Center for Magnetic Resonance Research, University of Illinois at Chicago, Chicago, IL, United States, 2Department of Electrical and Computer Engineering, University of Illinois at Chicago, Chicago, IL, United States, 3Research Resource Center, University of Illinois at Chicago, Chicago, IL, United States, 4Department of Radiology, Northwestern University, Chicago, IL, United States, 5Department of Computer Science, University of Illinois at Chicago, Chicago, IL, United States, 6Department of Bioengineering, University of Illinois at Chicago, Chicago, IL, United States, 7Department of Pathology, University of Illinois at Chicago, Chicago, IL, United States, 8Departments of Radiology, and Neurosurgery, University of Illinois at Chicago, Chicago, IL, United States
Synopsis
Tissue heterogeneity is an important consideration for diagnosing many diseases. Recently, a novel non-Gaussian diffusion model – continuous-time random-walk model (CTRW) – provided promising evidence indicating a possible link between voxel-level spatiotemporal diffusion heterogeneity and microscopic intravoxel tissue heterogeneity. Establishing a correlation between imaging-based and histology-based measurements, however, has been challenging because of the lack of efficient and subjective evaluation of tissue heterogeneity histologically. In this study, we applied a machine-learning algorithm to quantitatively determine microscopic tissue heterogeneity, enabling a correlation between intravoxel diffusion heterogeneity based on CTRW parameters and structural heterogeneity from histopathology.
Introduction
Tissue heterogeneity is one of the most important factors in disease diagnosis and treatment evaluation, particularly for cancer1-3. Tissue heterogeneity can arise from a variety of origins such as genetics, epigenetics, physiology, and pathology, all of which usually lead to structural intravoxel tissue heterogeneity at a specific scale, which is much smaller than the current spatial resolution of human MRI. A number of recent studies have shown that a novel diffusion-weighted imaging (DWI) technique based on a continuous-time random-walk (CTRW) model may provide a possible link between voxel-level anomalous diffusion parameters and microscopic intravoxel tissue heterogeneity4-8. Despite increasing evidence7,9-11, a rigorous correlation between the CTRW diffusion parameters and histology-based tissue heterogeneity has not been established. This is primarily because histopathologic assessment of tissue heterogeneity is hampered by lack of a consistent standard and labor intensity. In this study, we employ a machine-learning technique to determine microscopic tissue heterogeneity consistently and efficiently, allowing us to establish a correlation between diffusion-based and histology-based measurements of intravoxel tissue heterogeneity.Methods
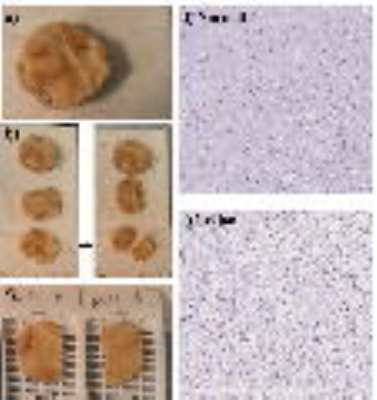
DWI Acquisition: A tissue specimen from a postmortem human brain with glioma-mimicking reactive gliosis was scanned on a 9.4T Agilent MRI scanner. The MRI protocol included anatomic T1 and T2 imaging, followed by DWI with 16 b-values from 0 to 4842 s/mm2 (TR/TE = 2000/28ms, slice thickness = 0.3mm, Δ = 18ms, δ = 2.5ms, FOV = 32mm×32mm, reconstruction matrix size = 128×128). Trace-weighted images were obtained to minimize the effect of diffusion anisotropy. Diffusion Image Analysis: The multi-b-value diffusion images were analyzed using a CTRW model8, $$S/S_0=E_α (-(bD_m )^β),$$ where Dm is an anomalous diffusion coefficient, Eα is a Mittag-Leffler function, and α and β are temporal and spatial diffusion heterogeneity parameters, respectively4-8. A nonlinear least-squares algorithm was used to obtain the CTRW parameters. Histological Staining: After MRI, the tissue specimen was prepared for histological staining as shown in Figs. 1a-1c. The sections for analysis included both normal and lesion tissues as shown in Figs. 1d and 1e. Machine-learning Classification Algorithm: The machine-learning algorithm was trained with histology images from 7 normal and 9 abnormal brain samples, which were labeled with varying degrees of microscopic heterogeneities by a senior neuropathologist. For each category, 50 prototype image regions (i.e., patches), each consisting of 1000×1000 independent pixels, were used. QuPath software was used to generate 33 statistical features characterizing nuclei and surrounding structures within each patch. To quantitatively classify brain tissue according to their heterogeneity levels, a random forest algorithm with Python's Scikit-Learn machine-learning package was employed. Model performance was validated using a 10-fold cross-validation. Establishing a Correlation between CTRW Parameters and Histology-based Metrics: The CTRW maps for the matched selections of the specimen were segmented into small patches similar to the technique used in the machine-learning algorithm. The CTRW parameter patches were classified as “homogeneous” or “heterogeneous” by the machine-learning algorithm. The statistical differences between the two categories in the mean CTRW parameters of the patches were evaluated by using a Mann-Whitney U test.Results
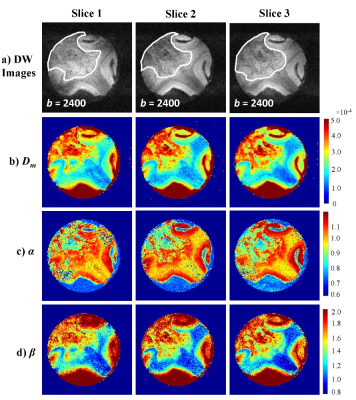
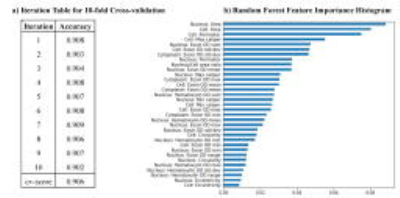
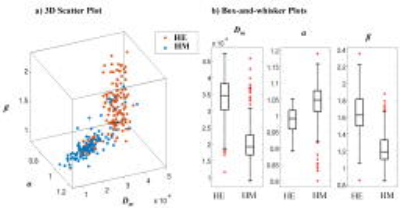
Figures 2a-2d display the diffusion images at b=2400 s/mm2 and the corresponding Dm, α, and β maps from three slices that correspond to the histology sections. The voxels in the lesion area (shown with white contours) exhibited significantly higher Dm and β, and lower α than those in the normal brain areas. The machine-learning-based classification of microscopic tissue heterogeneity resulted in a cross-validation score of 0.906, whose iterations are shown in Fig. 3a. The top three important features, according to the trained random forest classifier, included nucleus area, cell area, and cell perimeter as illustrated in Fig. 3b. A clear separation between the homogeneous and heterogeneous regions was observed in a three-dimensional scatter plot of CTRW parameters based on machine-learning classification (Fig. 4a). Box-and-whisker plots of the parameters (Fig. 4b) further illustrate this separation. All CTRW parameters yielded a statistically significant difference between homogeneous and heterogeneous patches (p-values<0.05).Discussion and Conclusion
Using a machine-learning algorithm for microscopic level tissue classification, we have shown that the CTRW model parameters can well-differentiate tissues with different structural heterogeneity at the sub-voxel level. A strong correlation was observed between intravoxel tissue heterogeneity measured by DWI and structural heterogeneity revealed by histopathology. With further validation, the CTRW model is expected to provide a new avenue for non-invasive in vivo characterization of microscopic tissue heterogeneity for normal and disease tissues.Acknowledgements
This work was supported in part by NIH 1S10RR028898. We thank Michael Flannery for technical assistance in tissue specimen preparation.References
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37.
- Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–64.
- Fletcher CDM. The evolving classificiation of soft tissue tumors – an update based on the new 2013 WHO classification. Histopathology. 2014;64(1):2-11.
- Magin RL, Abdullah O, Baleanu D, et al. Anomalous diffusion expressed through fractional order differential operators in the Bloch-Torrey equation. J Magn Reson Im. 2008;190:255–270.
- Zhou XJ, Gao Q, Abdullah O, Magin RL. Studies of anomalous diffusion in the human brain using fractional order calculus. Magn Reson Med. 2010;63:562–569.
- Ingo C, Magin RL, Colon-Perez L, et al. On random walks and entropy in diffusion-weighted magnetic resonance imaging studies of neural tissue. Magn Reson Med. 2014;71(2):617–27.
- Sui Y, He W, Damen FW, et al. Differentiation of low- and high-grade pediatric brain tumors with high b-value diffusion weighted MR imaging and a fractional order calculus model. Radiology. 2015;277(2):489–496.
- Karaman MM, Sui Y, Wang H, et al. Differentiating low- and high-grade pediatric brain tumors using a continuous-time random-walk diffusion model at high b-values. Magn Reson Med. 2015; 76:1149-1157.
- Sui Y, Xiong Y, Xie KL, et al. Differentiation of low- and high-grade gliomas using high b-value diffusion imaging with a non-Gaussian diffusion model. Am J Neuroradiol. 2016; 37:1643-1649.
- Tang L, Sui Y, Zhong Z, et al. Non-Gaussian diffusion imaging with a fractional order calculus model to predict response of gastrointestinal stromal tumor to second-line sunitinib therapy. Magn Reson Med. 2017.
- Liu G, Xie K, Sui Y, et al. Diffusion-weighted MR imaging of prostate with a fractional order calculus model. In: Proceedings of the 21st Annual Meeting of ISMRM 2013;1783.
Figures