3103
Estimation of Extracellular Matrix Diffusion Properties in Decellularized Porcine Myocardium from DTI1Univeristy of Illinois at Urbana Champaign, Urbana, IL, United States, 2Illinois Institute of Technology, Chicago, IL, United States
Synopsis
We present a method to estimate microstructural parameters of a decellularized pig myocardium using a two-compartment exchange model. We also show that the estimated parameters are in good agreement with other values found in the literature.
Introduction
Myocardium decellularization has been established as a method to produce intact three-dimensional scaffolds for bioartificial hearts,1 and patches for cardiac repair.2,3 The process can be used as a potential tool to study the structure of the extracellular matrix (ECM). Mass transport in the ECM is also a factor in MR imaging of the heart, and knowledge of the histoarchitecture is important in interpreting cardiac DTI measurements.3,4 Like skeletal muscle, the myocardium is characterized by a hierarchical organization of collagenous ECM. Bundles of myocytes registered by endomysial collagen are organized into twisting layers in a characteristic transmural direction. The perimysium tethers these layers and features large convoluted collagen cords aligned approximately with the myocyte axis. A range of various transmural connections is possible within such a hierarchy of muscle layers.5 The complex 3-D structure of the myocardium and its spatial discontinuity makes modeling the ECM microstructure a challenge. The transverse isotropy assumption is also not necessarily valid due to the sheet like structure of the ECM in the heart. All of these factors complicate the requirements needed to model diffusion in the cardiac muscle, compared to skeletal muscle. We present a modified Karger model which allows for exchange between two diffusion pools and also incorporates restrictions to the diffusion coefficient due to microstructural barriers such as the sarcolemma membrane. By fitting this model to DTI data from a phantom consisting of decellularized porcine myocardium we are able to estimate microstructure parameters of the phantom.Methods
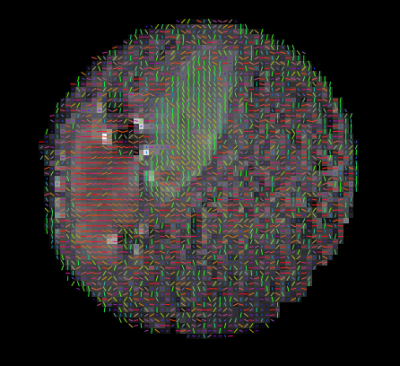
Two sections of decellularized porcine myocardium were placed in a lipid-gel mixture and imaged using a Siemens 3T (TRIO) scanner with an epi_mddw_30 DTI sequence consisting of 30 gradient directions at b = 1000 s/mm2 and TE/TR = 95/3500 ms. Data was post processed with FSL to determine the diffusion tensor.6 Additionally, a numerical model based on the Karger model of two compartment exchange7 was used to fit parameters. The model consists of an intracellular and extracellular pool of spins with exchange between them. Each pool has an anisotropic diffusion tensor with the intracellular pool modeled as infinitely long cylinders with an axial (Din,a) and transverse (Din,t) diffusion coefficient while the extracellular pool reflects the model of the ECM proposed by Tseng et al4 with an anisotropic diffusion tensor with the largest diffusion coefficient (Dex,1) related to diffusion within the ECM in the axial direction, the secondary diffusion coefficient (Dex,2) related to diffusion parallel to layers of myocytes and ECM and the tertiary diffusion coefficient (Dex,3) is perpendicular to these sheets with restriction caused by the myocyte membranes. These 5 diffusion coefficients are combined with an intracellular exchange time and an intracellular inclusion fraction to provide 7 free parameters which were fit to the 30 direction data at each voxel. The model was fit to each section independently using two manually identified FOIs. Fitted parameters were filtered such that any voxel with a fitted parameter 3 standard deviations beyond the mean was excluded.Results
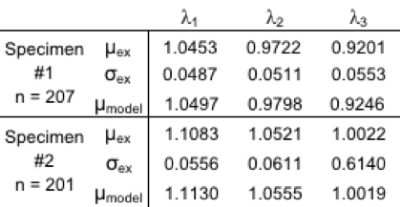
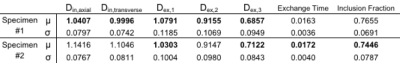
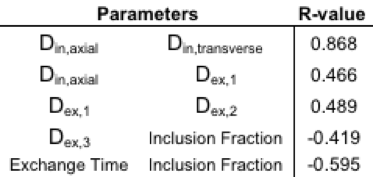
The eigenvalues from the phantom were normally distributed and comparison was made between the mean of the phantom’s eigenvalues and the numerically predicted eigenvalues using the mean of the 7 fitted parameters (Table 1). 9 of the 14 fitted parameter sets were found to be normally distributed with the other five sets demonstrating skewness. The mean and standard deviations of the fitted parameters are shown in Table 2. Finally, correlation tests were performed to compare how the fitted parameters correlated with each other. Correlation was found among 5 parameter sets as shown in Table 3.Discussion
The fitted parameters are realistic and fit literature values for packing fraction.8The difference in Din is suspected to be due to different amounts of decellularization between the two specimens. The exchange time relates to cell sizes ~20 mm with a sarcolemma permeability of ~500 mm/s, this diameter of 20 mm fits literature values2,9,10 while permeability is higher than published values of sarcolemma permeability (10-400 mm/s),11 but due to the decellularization process it is plausible that the membrane also degraded leading to higher permeability.Conclusion
We have developed a model that fits structural parameters to the heart while incorporating cardiac microstructure and exchange between ECM and myocytes domains. This model was fitted to DTI data from phantom which consisted of two sections of decellularized pig heart and produced results that match literature values with correlations between parameters which are well explained by the model. These results suggest that the two-compartment model can be used to estimate microstructural parameters of the myocardium.Acknowledgements
Funded for this work was provided by a NSF Graduate Research FellowshipReferences
1 Ott, Harald C., et al. "Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart." Nature medicine14.2 (2008): 213-221.
2 Wang, Bo, et al. "Fabrication of cardiac patch with decellularized porcine myocardial scaffold and bone marrow mononuclear cells." Journal of biomedical materials research Part A 94.4 (2010): 1100-1110.
3 Saeed, Maythem, et al. "Cardiac MR imaging: current status and future direction." Cardiovascular diagnosis and therapy 5.4 (2015): 290.
4 Tseng, Wen‐Yih I., et al. "Diffusion tensor MRI of myocardial fibers and sheets: correspondence with visible cut‐face texture." Journal of Magnetic Resonance Imaging 17.1 (2003): 31-42.
5 LeGrice, Ian, Adéle Pope, and Bruce Smaill. "The architecture of the heart: myocyte organization and the cardiac extracellular matrix." DEVELOPMENTS IN CARDIOVASCULAR MEDICINE253 (2005): 3.
6 Jenkinson, Mark, et al. "Fsl." Neuroimage 62.2 (2012): 782-790.
7 Stanisz, Greg J., et al. "An analytical model of restricted diffusion in bovine optic nerve." Magnetic Resonance in Medicine 37.1 (1997): 103-111.
8 Zhou, Pingzhu, and William T. Pu. "Recounting cardiac cellular composition." (2016): 368-370.
9 Kiczak, Liliana, et al. "Sex differences in porcine left ventricular myocardial remodeling due to right ventricular pacing." Biology of sex differences 6.1 (2015): 32.
10 Wang, Bo, et al. "Structural and biomechanical characterizations of porcine myocardial extracellular matrix." Journal of Materials Science: Materials in Medicine 23.8 (2012): 1835-1847.
11 Fieremans, Els, et al. "In vivo measurement of membrane permeability and myofiber size in human muscle using time‐dependent diffusion tensor imaging and the random permeable barrier model." NMR in Biomedicine 30.3 (2017).
Figures