3078
Location-wise Harmonization along White Matter Tracts on Neonatal Brain from Multiple Acquisition Protocols1Department of Diagnostic Radiology, the First Affiliated Hospital of Xi’an Jiaotong University, Xi'an, China, 2Department of Biomedical Engineering, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an, China, 3Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China
Synopsis
In this work, we proposed white matter location-wise harmonization of DTI measurement on neonatal brain based on tract quantification from multiple acquisition protocols. A total of 120 term neonates examined by two DTI acquisition protocols, including 38 neonates with punctate white matter lesions and 82 without MRI abnormalities, were enrolled. Two methods, Scaling and ComBat, were adopted, each carried out also on global-wise and ROI-wise in addition to location-wise. We also proposed an evaluation framework to systematically compare the adopted harmonization approaches, which suggests the way to select the utmost harmonization level and the specific method.
Purpose
Diffusion tensor imaging (DTI) is widely used in neonatal brain study, where tract quantification reveals new insight1. Large samples are necessary to investigate microstructural information of white matter development or lesions. Long-term or multi-center studies are common strategies to enlarge samples and increase statistical power2. However, both strategies suffer from acquisition protocols variation, adversely affecting accuracy of data joint analysis. Several harmonization approaches2-4 were neither applied to clinical patient data nor tract quantification based. This work aimed to harmonize DTI measurement along white matter tracts across multiple acquisition protocols, with selecting the utmost clinically practical method applied to neonatal punctate white matter lesions (PWMLs) study, by comparing the performance of various harmonization approaches.Methods
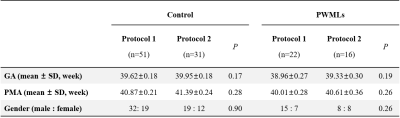
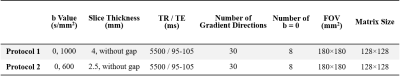
Subjects and Data Acquisition This study was approved by the local institutional review board. Parents were informed of the goals and risks of MR scan and signed the consent form. Inclusion criteria was evidence of punctate lesions in cerebral white matter on T1WI and T2WI. We excluded neonates with congenital malformations of central nervous system, major destructive white matter lesions, gray matter lesions, metabolic disorders, hydrocephalus and infections. Gender, gestational age (GA) and postmenstrual age (PMA) matched neonates without MRI abnormalities and episodes were enrolled as controls (Table 1). MRI data were acquired for clinical examination, with oral chloral hydrate sedation and neonate monitoring performed strictly following guidelines. Three-dimensional fast spoiled gradient-recalled echo T1WI, fast spin echo T2WI and DTI by single-shot echo planar imaging were performed on a 3.0 T scanner (General Electric, Signa HDXT, WI, USA) with an eight-channel head coil. Two DTI acquisition protocols were adopted, with b values and slice thickness varied (Tabel 2).
Harmonization Two methods, Scaling3 and ComBat4, were adapted to global-wise, ROI-wise as well as location-wise tract quantification based approaches, using harmonization factors extracted from mean FA of whole brain in the control groups, interested white matter tracts and tract location respectively. FA calculation and normalization to neonatal template5 were performed in FSL (FMRIB software library, University of Oxford, UK). We quantified FA on 100 equally spaced locations1 along right corticospinal tract (rCST) and splenium of the corpus callosum (SCC), using tract probabilistic maps. The selected tracts showed vulnerability to PWMLs in a previous study6.
Evaluation Framework Above approaches were compared in our proposed evaluation framework. The harmonization is successful if: 1) it removes inter-protocol differences induced by acquisition parameters, 2) meanwhile, preserves PWMLs-related biological variability. We set two PWMLs-related variability references: comparison between PWMLs 1 and Control 1, comparison between PWMLs 2 and Control 2, both before harmonization. Wilcoxon signed rank test was performed to evaluate differences in GA, PMA, FA values across protocols and PWMLs-related variability. Chi-square test was performed to evaluate inter-protocol gender difference and consistency of PWMLs-related variability before and after harmonization.
Results and Discussion
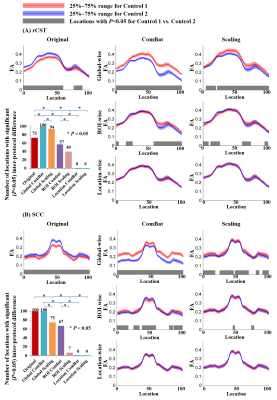
Fig. 1 emphasizes the importance of harmonization and suggests the selection of location-wise approaches. Before harmonization, significant inter-protocol differences of FA values exited along both tracts. The efficacy of removing inter-protocol differences differs across harmonization levels and also methods, with global-wise harmonization performing worst while location-wise Scaling and ComBat performing perfectly.
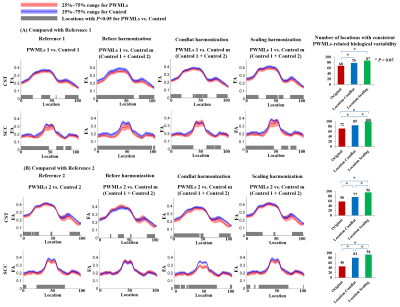
Fig. 2 reveals the better performance of location-wise Scaling on preserving PWMLs-related biological variability after harmonization. Before harmonization, when merging the data of control groups, the consistency of PWMLs-related variability along both tracts was quite low. Location-wise harmonization improves the consistency significantly. Particularly, location-wise Scaling improved the consistency more significantly (P<0.05) than ComBat.
In this work, we initially harmonized DTI measurement on data from clinical patients: neonates with PWMLs. Additionally, a proposed evaluation framework used the clinically interest PWMLs-related biological variability, to validate applicability of harmonization approaches on patient data. The feasibility that harmonization factors can be extracted from control groups is based on the following assumption: for the two demographics-matched control groups, inter-group differences of FA values are only caused by acquisition parameter differences2. Location-wise harmonization has the advantage of not obscuring detailed information of each tract location, as well as avoiding inaccuracy of coregistration in voxel-wise approach2. Smoothing effect of ComBat4 may lower its accuracy on regional FA abnormalities caused by PWMLs than Scaling.
Conclusion
This study shows that white matter location-wise harmonization improves the efficacy of removing unwanted inter-protocol differences. Particularly, location-wise Scaling method shows the best stability of preserving PWMLs-related biological variability as well as removing inter-protocol differences. Both the proposed evaluation frame work and selected harmonization method have the potential of extended clinical application in multi-center studies.Acknowledgements
This study was supported by the National Key Research and Development Program of China (2016YFC0100300), National Natural Science Foundation of China (No. 81471631, 81771810 and 51706178), the 2011 New Century Excellent Talent Support Plan of the Ministry of Education, China (NCET-11-0438) and the Clinical Research Award of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF-CRF-2015-004).References
[1] Yeatman J D, Dougherty R F, Myall N J, et al. Tract Profiles of White Matter Properties: Automating Fiber-Tract Quantification[J]. Plos One, 2012, 7(11): e49790.
[2] Mirzaalian H, Ning L, Savadjiev P, et al. Inter-site and inter-scanner diffusion MRI data harmonization[J]. Neuroimage, 2016, 135: 311-323.
[3] Pohl K M, Sullivan E V, Rohlfing T, et al. Harmonizing DTI measurements across scanners to examine the development of white matter microstructure in 803 adolescents of the NCANDA study[J]. Neuroimage, 2016, 130: 194-213.
[4] Fortin J P, Parker D, Tunç B, et al. Harmonization of multi-site diffusion tensor imaging data[J]. Neuroimage, 2017, 161: 149-170.
[5] Oishi K, Mori S, Donohue P K, et al. Multi-contrast human neonatal brain atlas: Application to normal neonate development analysis[J]. Neuroimage, 2011, 56(1): 8-20.
[6] Li X, Gao J, Wang M, et al. Characterization of Extensive Microstructural Variations Associated with Punctate White Matter Lesions in Preterm Neonates[J]. American Journal of Neuroradiology, 2017, 38(6): 1228-1234.
Figures