3077
Feasibility and benefits of 3-tissue constrained spherical deconvolution for studying the brains of babies1The Florey Institute of Neuroscience and Mental Health, Melbourne, Australia, 2Department of Audiology and Speech Pathology, University of Melbourne, Melbourne, Australia, 3The HEARing Cooperative Research Centre, Melbourne, Australia, 4The Florey Department of Neuroscience, University of Melbourne, Melbourne, Australia
Synopsis
When studying white matter in baby brains with diffusion-weighted imaging, we face a range of challenges, including larger water-content, lower anisotropy, differentiated maturation and a (relatively) larger proportion of the brain being comprised of grey matter. We attempt to apply single-tissue, 2-tissue and 3-tissue constrained spherical deconvolution (CSD) to single-shell data of two 5 month old babies. 3-tissue CSD still worked successfully. The nature of benefits was in line with those obtained previously in adults, but they were greater in the babies, mostly due to a much larger presence of GM-like tissue.
Purpose
Imaging the baby brain brings a range of challenges, including partial voluming and (often) practical limitations on scan time. Diffusion-weighted imaging (DWI) in particular may be challenged in these areas; but when studying the white matter (WM), even further complications arise due to larger water-content, much lower anisotropy and strongly differentiated maturation in the developing brain[1], and a (relatively) larger proportion of the baby brain being comprised of grey matter (GM)[2].
Constrained spherical deconvolution (CSD)[3] models the DWI-signal using only a single-fibre WM response function, and hence may be too simplistic to take on the aforementioned challenges. Multi-shell multi-tissue CSD (MSMT-CSD)[4] and single-shell 3-tissue CSD (SS3T-CSD)[5] resolve additional GM-like and CSF-like compartments by adding GM/CSF response functions to the model. Multi-tissue methods have led to good results in adult brains, but whether they perform equally well in babies remains unknown.
In this work we attempt to analyse previously acquired DWI-data of 2 babies (both acquired using a typical single-shell, high b-value, high-angular resolution protocol) using different (multi-tissue) CSD strategies.
Data and Preprocessing
- Baby 1: DWI-data from a 5 month old baby (normally developing) were acquired using a Siemens 3T Trio scanner: voxel size 2.5×2.5×2.5mm³, b=3000s/mm², 60 gradient directions, 8 b=0 images.
- Baby 2: DWI-data from a 5 month old baby (with unilateral hearing loss due to hypoplasia of the auditory nerve (right side), but otherwise normally developing) were acquired using a Siemens 3T Skyra scanner: voxel size 2.3×2.3×2.3mm³, b=2800s/mm², 64 gradient directions, 8 b=0 images; multiband factor 3. This protocol was repeated twice; all data were combined into a single DWI-dataset.
All data were denoised[6], corrected for Gibbs-ringing[7], distortions/motion[8,9], bias-fields[10].
Methods
Response functions were estimated from the DWI-data using an unsupervised method[11]. We did have to tune a single parameter in this method: an FA threshold responsible for initial (crude) WM segmentation is by default set to 0.2[11]. We lowered it to 0.1, in line with lower overall FA in the baby-brains[1]. The method performed as expected otherwise, selecting appropriate single-fibre WM, GM and CSF voxels for response function calibration.
Next, both datasets were up-sampled to 1.2×1.2×1.2mm³ and 3 different CSD strategies were performed:
- single-tissue: using (single-shell) CSD[3], the WM response is deconvolved from the high-b-valued shell (ignoring b=0 data), to obtain the WM-FOD
- 2-tissue: using MSMT-CSD[4], the WM and CSF responses are deconvolved from all data (including b=0), to obtain the WM-FOD and CSF compartment
- 3-tissue: using SS3T-CSD[5], the WM, GM and CSF responses are deconvolved from all data, to obtain the WM-FOD, GM and CSF compartments
Finally, probabilistic tractography[12] was run, guided by each CSD strategy's WM-FOD output.
Results and Discussion
Fig.1–2 show an overview of results obtained by applying the different CSD strategies to both datasets. While adding a CSF-like compartment to the model by 2-tissue CSD slightly reduced the overestimation of WM-like signal (by single-tissue CSD), the impact was relatively limited. A much greater effect was achieved by adding a GM-like compartment to the model via 3-tissue CSD. The method is still able to successfully tease apart realistic contributions from all 3 tissue types, which we hypothesised could have been a problematic challenge[16]. Not just the WM-like signal was still overestimated by 2-tissue CSD, but CSF-like signal as well. This could impact assessment of the free-water content in the brain, which is a typical feature of interest in developing white matter.
Fig.3 presents the WM-FODs from each CSD strategy. The impact of (not) overestimating the WM is clear: accounting for GM-like signal using 3-tissue CSD markedly "cleans up" the WM-FODs in the cortical area, revealing well-known radial organisation[15]. As expected, this has a strong impact on the quality of tractography outcomes informed/guided directly by these WM-FODs (Fig.4–5).
Finally, we note that others have attempted to apply multi-tissue CSD to neonatal brains[16]. Due to ambiguities when differentiating WM and GM, they chose to apply only 2-tissue CSD. While our data doesn't reflect the challenges neonatal data additionally brings, it does present issues that will remain when not explicitly accounting for GM-like signal. These issues are expected to be even greater in neonates, given a proportionally even larger presence of GM[2]. The work in [16] also used multi-shell data, which may (paradoxically) introduce other problems[17] when using a model that is too simplistic with respect to the data.
Conclusion
We were successfully able to apply 3-tissue CSD in brains of 5 month old babies. The nature of the benefits was in line with those obtained previously in adults, but they were greater in the babies, mostly due to a much larger presence of GM-like tissue.Acknowledgements
No acknowledgement found.References
[1] Yu Q, Kang H, Peng Q, Ouyang M, Slinger M, Peng Y, Fang F, Huang H. Differentiated maturation of white matter tracts in early developing brain aged 0-3 years. Proc ISMRM 2017;25:1267.
[2] Groeschel S, Vollmer B, King MD, Connelly A. Developmental changes in cerebral grey and white matter volume from infancy to adulthood. Int J Dev Neurosci 2010;28(6):481-489.
[3] Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. NeuroImage 2007;35(4):1459-1472.
[4] Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage 2014;103:411-426.
[5] Dhollander T, Connelly A. A novel iterative approach to reap the benefits of multi-tissue CSD from just single-shell (+b=0) diffusion MRI data. Proc ISMRM 2016;24:3010.
[6] Veraart J, Fieremans E, Novikov DS. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med 2016;76(5):1582-1593.
[7] Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med 2016;76(5):1574-1581.
[8] Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 2003;20(2):870-888.
[9] Andersson JLR, Xu J, Yacoub E, Auerbach E, Moeller S, Ugurbil K. A comprehensive Gaussian Process framework for correcting distortions and movements in diffusion images. Proc ISMRM 2012;20:2426.
[10] Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: Improved N3 bias correction. IEEE TMI 2010;29(6):1310-1320.
[11] Dhollander T, Raffelt D, Connelly A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. Proc ISMRM Workshop on Breaking the Barriers of Diffusion MRI 2016:5.
[12] Tournier JD, Calamante F, Connelly A. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. Proc ISMRM 2010;18:1670.
[13] Dhollander T, Smith RE, Tournier JD, Jeurissen B, Connelly A. Time to move on: an FOD-based DEC map to replace DTI's trademark DEC FA. Proc ISMRM 2015;23:1027.
[14] Dhollander T, Raffelt D, Connelly A. Towards interpretation of 3-tissue constrained spherical deconvolution results in pathology. Proc ISMRM 2017;25:1815.
[15] McKinstry RC, Mathur A, Miller JH, Ozcan A, Snyder AZ, Schefft GL, Almli CR, Shiran SI, Conturo TE, Neil JJ. Radial organization of developing preterm human cerebral cortex revealed by non-invasive water diffusion anisotropy MRI. Cereb Cortex 2002;12(12):1237-1243.
[16] Pietsch M, Hutter J, Price A, Murgasova MK, Hughes E, Steinweg J, Tusor N, Andersson J, Bastiani M, Sotiropoulos S, Hajnal JV, Tournier JD. Multi-shell neonatal brain HARDI template. Proc ISMRM 2017;25:1268.
[17] Jbabdi S, Sotiropoulos SN, Savio AM, Graña M, Behrens TE. Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magn Reson Med 2012;68(6):1846-1855.
Figures
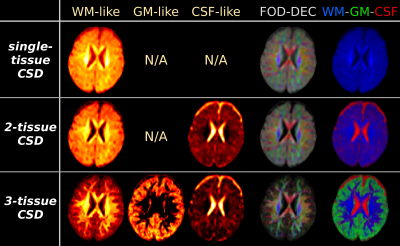
Fig.1: results from the CSD strategies, applied to "baby 1".
All WM-like, GM-like and CSF-like signal compartment maps use the same windowing/scale. The WM-FOD based directionally-encoded colours (FOD-DEC[13]) are derived from the WM-FOD itself (red/green/blue = mediolateral, anteroposterior, superoinferior), weighted by WM-like compartment intensity. The WM-GM-CSF tissue map combines the WM/GM/CSF-like signal compartment maps in a single visualisation for overview (red/green/blue = CSF/GM/WM).
Note the biggest impact on the WM-like compartment is achieved by the addition of a GM-like compartment in the model (via 3-tissue CSD). This also has an impact on the estimated CSF-like compartment (relative to 2-tissue CSD).
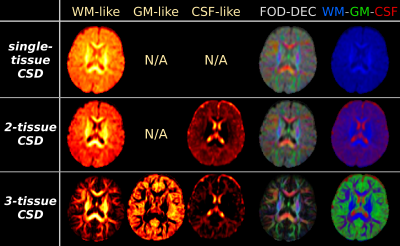
Fig.2: results from the CSD strategies, applied to "baby 2". Equivalent maps to Fig.1 are shown.
As in Fig.1, the biggest impact on the WM-like compartment is achieved by adding a GM-like compartment to the model, via 3-tissue CSD. A very large portion of voxels shows GM-like signals. Compared to 2-tissue CSD, this impacts both WM-like and CSF-like compartments, which are both overestimated by 2-tissue CSD. Note CSF-like signal also models interstitial fluid (which is differentially present in the WM during early development), and GM-like signal may also model excess extra-axonal presence of glial cells or other non-axonal content[14].
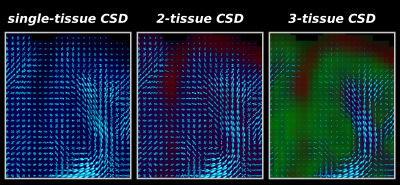
Fig.3: Zoomed frontal region of the same axial slice as shown in Fig.2, showing WM-FODs from each CSD strategy superimposed on the tissue map (darkened).
2-tissue CSD filters out CSF-like signal, which is mostly present outside the brain parenchyma, and to a lesser extent in some WM regions (see also Fig.2), but the impact on the quality of the WM-FODs is fairly limited (relative to single-tissue CSD).
3-tissue CSD additionally filters out GM-like signal, which still had a large contribution to the WM-FOD in 2-tissue CSD. This drastically improves WM-FOD quality in GM, and reveals radial organisation of the cortex[15].
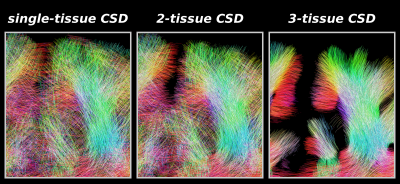
Fig.4: Same region as shown in Fig.3, showing tractography results as guided by the WM-FODs from each CSD strategy.
As expected from the quality of the WM-FODs (Fig.3), improvement of 2-tissue CSD relative to single-tissue CSD is mostly outside / on the edge of the brain parenchyma: fewer false positive streamlines connect both hemispheres across CSF.
3-tissue CSD guided tractography shows marked improvement: more coherent streamlines that penetrate into the cortex in line with the radial organisation observed in Fig.3. Tractography is still limited, failing to make certain sharp turns into the cortex, even though local WM-FODs show proper structure.
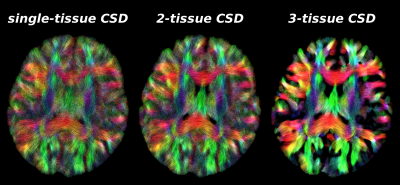
Fig.5: Same axial slice as shown in Fig.2, showing tractography results as guided by the WM-FODs from each CSD strategy.
Similar observations as in Fig.4 hold across the brain. 2-tissue CSD reduces some false positive streamlines that otherwise travel across CSF areas in the single-tissue CSD result, and improves streamline coherence in some WM areas.
3-tissue CSD shows widespread improvements in coherence of streamlines and reduction of false positives. This is not surprising, given the amount of GM (partial voluming) in the baby brain[2]. Remaining limitations are due to streamline tractography failing to make certain sharp turns into the cortex.