3075
2-year-old human brain DTI atlas with comprehensive gray and white matter labels1Shandong University School of Medicine, Jinan, China, 2The Children’s Hospital of Philadelphia, Philadelphia, PA, United States, 3Beijing Children’s Hospital Affiliated to Capital Medical University, Beijing, China, 4Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
Synopsis
2-year-old, marking the end of infancy, is critical for understanding not only precisely organized normal brain development but also serving as clinical anatomical references for neurodevelopmental disorders such as autism. The 2-year-old brain labels transformed from adult atlases lead to relatively large offsets due to dramatic and nonlinear neuroanatomical differences of the brains between these two populations. With DTI data from nineteen healthy 2-year-old subjects, we created a 2-year-old brain DTI atlas with comprehensive labels of 124 gray and white matter structures. The test results suggested the established atlas can be applied to label 2-year-old brain images automatically and accurately.
Target audience
Pediatric neuroradiologists, psychiatrists and neurologists.Purpose
Dramatic brain structural changes take place in the first 2 years of life which is also characterized by emergence of neurodevelopmental disorders such as autism [1]. Atlas-based automated structure labeling is useful for analyzing functional and structural neuroimaging data. Due to the relative large and nonlinear neuroanatomical differences between pediatric and adult brains, the pediatric brain parcellation using adult atlases can lead to significant offsets of labeled structures. Hence, the age-specific 2-year-old atlas with comprehensive gray matter (GM) and white matter (WM) labels is needed. In this study, with diffusion tensor imaging (DTI) data of nineteen healthy 2-year-old subjects, we created a 2-year-old brain DTI atlas with comprehensive labels of 124 GM and WM structures. Integrated with appropriate registration methods, the atlas can be used for automatic labeling of given 2-year-old brain images with high accuracy and small offsets of labeling.Material and Methods
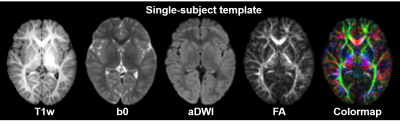
Subjects, data acquisition and preprocessing: Nineteen toddlers with postnatal age of 20 to 28 months participated in this study. T1w images and DTI data were acquired on a 3T Philips Achieva System. The DTI imaging parameters were: TE = 100ms, TR = 9300ms, in-plane field of view = 256 x 256mm2, in-plane imaging resolution = 128 x 128mm2, slice thickness = 2mm, slice number = 70, 30 independent diffusion encoding directions, b-value = 1000s/mm2, repetition = 2. To obtain co-registered T1w and DTI images of each subject, affine and large deformation diffeomorphic metric mapping transformations [2] were used to warp b0 images to the contrast-reversed T1w images using DiffeoMap (mristudio.org). The resultant registration matrix was applied to transfer diffusion tensor [3]. After these procedures, the co-registered T1w and DTI images were created, with a 1.0mm isotropic resolution. Single-subject and population-averaged template: Single-subject template (Fig.1) was established using Diffeomap, following the steps elaborated in the literature [4]. To create the population-averaged template, we registered the T1w images of all participating toddlers to a representative single subject using AIR transformation. Establishment of the comprehensive gray and white matter brain atlas: Comprehensive labels of 124 GM and WM structures were obtained with manual delineation on axial planes of the single-subject template with ROIEditor (mristudio.org), followed by adjustment in coronal and sagittal planes using available atlases [4-5] as references. The deep WM and subcortical GM nuclei were labeled with DTI orientation-encoded colormaps and T1w images, respectively. The cerebral cortex and superficial WM structures were delineated based on the sulcal and gyral patterns on the T1w images followed by the modifications based on 3D reconstructed labeled structures using Amira (FEI software, Hillsboro, OR). Test of the automated labeling using the established atlas: Five subjects were selected for testing automated GM and WM labeling using the established atlas. The automated labels were tested against manual labels with measurements of Dice coefficients and L1 errors.Results
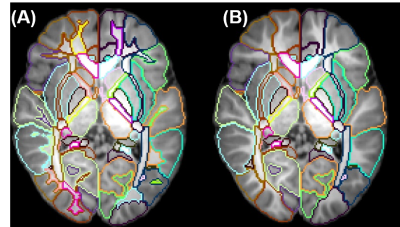
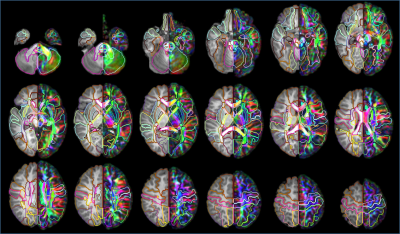
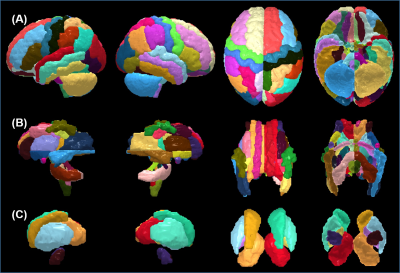
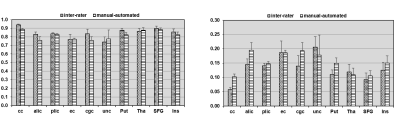
Two types of atlases: Two types of atlases were generated with superficial WM separated in Type-I atlas (Fig 2A) and superficial WM integrated with associated cortical labeling in Type-II atlas (Fig 2B), with Type-II atlas demonstrated in details in this abstract. 2- and 3-dimensional comprehensive gray and white matter labels: Fig 3 shows the Type-II atlas on the T1w images and orientation-encoded colormaps in axial planes with comprehensive GM and WM labels of 52 cerebral cortical structures, 40 deep cerebral WM tracts, 10 subcortical GM structures and 22 brainstem and cerebellar structures. The 3D reconstructed labeled cerebral cortex, deep WM tracts, and subcortical GM are demonstrated in Fig 4A, 4B and 4C, respectively. Test of the automated labeling using the established atlas: As shown in Fig 5, the Dice coefficients between automated and manual delineation were over 0.8 in most structures, indicating almost perfect automated labeling. The Dice coeffiecients were more than 0.73 in cingulate and uncinate fasciculus, indicating registration accuracy even for the irregular anatomical structures. The misclassification analysis using L1 error indicate no statistically significant difference of misclassification between automated labeling and manual inter-rater variability.Discussion and Conclusions
We established a 2-year-old DTI atlas with comprehensive GM and WM labels of 52 cerebral cortical structures, 40 deep cerebral WM tracts, 10 subcortical GM structures and 22 brainstem and cerebellar structures. The test results suggested that the established atlas can be applied to label a given 2-year-old brain images automatically and accurately. The established 2-year-old DTI atlas may be used for understanding not only normal brain development but also serving as clinical anatomical references for neurodevelopmental disorders such as autism.Acknowledgements
This study is funded by NIH MH092535, MH092535-S1 and HD086984References
[1] Hazlett et al. (2017). Nature, 542:348. [2] Miller et al. (2002). Annual Review of Biomedical Engineering, 4:375. [3] Xu et al. (2003). Magnetic Resonance in Medicinde, 50:175. [4] Oishi et al. (2011). Neuroimage, 56(1):8. [5] Oishi et al. (2009). Neuroimage, 46(2):486.Figures