3045
Stem Cell Tracking Using Effective Self-Assembled Peptide-Modified Superparamagnetic Nanoparticles1Huaxi MR Research Center (HMRRC), Sichuan University, ChengDu, China, 2Department of Radiology, Molecular Imaging Program at Stanford (MIPS), Stanford University, Stanford, CA, United States
Synopsis
In cell therapies and regeneration medicine, superparamagnetic iron oxide nanoparticles (SPIONs) have been developed as excellent magnetic resonance imaging (MRI) contrast agents for stem cell labeling and tracking due to their biocompatibility. Here, we designed a self-assembled peptide amphiphile (PA) replace the transfection agents. This PA was conjugated to the surfaces of SPIONs to label rat mesenchymal stem cells (MSCs), which enhanced the contrast and labeling effects. The labeled cells showed that peptide-SPIONs had improved internalization, efficiency and T2-weight relaxivity and were nontoxic to the MSCs. The results demonstrated that these self-assembled peptide-modified SPIONs are potential candidates to label MSCs for tracking stem cells using MRI in vivo.
INTRODUCTION
The aim of this study was to design a self-assembled peptide modified superparamagnetic iron oxide nanoparticles (SPIONs) as excellent magnetic resonance imaging (MRI) contrast agents for stem cell labeling and tracking1. As the potential MR contrast agent, SPIONs should maintain excellent colloidal stability under physiological conditions, and the mainly effect is performed cell tracking in MR imaging. But the SPIONs uptake by stem cells is relatively inefficient, previous research has shown that SPIONs with surface modifications can increase the uptake efficiency of stem cells2. Different from many required positively charged transfection agents, peptides have good biocompatibility and biosafety. Besides, the high molecular weight increases the surface charge and hydrodynamic size of nanoparticles, which can improve the cell internalization. The self-assembling pwptide-amphiphile nanofibers (PAs) can be easily be modified or coupled with characterized chemical compounds, and peptides have been utilized for drug delivery and tissue regeneration due to their biocompatibility, including to modify MR agents3,4. The purpose of this study was to design the self-assembling lipopeptide modified SPIONs can facilitate MSCs labeling and also enhance the r2 relaxitivity value.METHODS
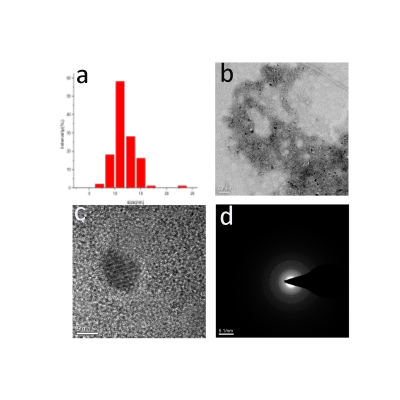
In this study, the synthesis of designed self-assembling peptides was performed by solid-phase synthesis methods. The iron oxide nanoparticles were prepared via polyol synthesis. And the peptide conjugation procedure was performed through the EDC method. Formation of the peptide-SPIONs was characterized by the means of dynamic light scattering (DLS), transmission electron microscopy (TEM), Fourier transform infrared (FTIR) spectroscopy and energy-dispersive X-ray spectroscopy (EDS) coupled to transmission electron microscopy (SEM). Rat MSCs were isolated form the bone marrow of SD rats. The relativity characterization of nanoparticles and the MR imaging of the labeled cells were acquired using a 7.0 Tesla MR scanner. Iron uptake was evaluated with Prussian blue staining and TEM micrograph. To test the safety of peptide-SPIONs, the cell proliferation rate was evaluated using the CCK-8 assay; flow cytometry was used to investigated cellular apoptosis and cell cycle distribution; alizarin red staining and oil red O staining assays are used to determine whether the differentiation potential of MSCs was maintained following labeling.RESULTS
The synthesis process for the peptide-SPIONs is presented in Scheme 1(Figure 1). Representative TEM micrographs of the peptide-modified nanoparticles had monodispersity and a uniform size (Figure 2). The FITR spectra results demonstrated that the Fe3O4 particles were successfully conjugated to lipopeptides. The r2 values of DMSA-SPIONs and peptide-SPIONs were 168.4 s-1/mM and 194.2 s-1/mM, respectively, which were significantly different between the two nanoparticles (p < 0.05) (Figure 3). The statistical analysis of the PB stain indicated that the peptide-modified SPIONs could improve the targeting efficiency and the rate of positive labeling up to 95%, which was clearly higher than the values for DMSA-SPIONs (Figure 4). The cell toxicity assays all determined that peptide-SPIONs are non-toxic effects in labeling progress.DISCUSSION
Peptide-SPIONs labeling MSCs has higher relaxation rate. This observation may be explained the higher relaxivity of the modified nanoparticles used and the higher uptake efficiency of peptide-modified nanoparticles into the cells. The high molecular weight effectively promoted the internalization of the nanoparticles by MSCs and prolonged their circulation time within the cells.CONCLUSION
The labeled cells showed that peptide-SPIONs had improved internalization, efficiency and T2-weight relaxivity and were nontoxic to the MSCs. The results demonstrated that these self-assembled peptide-modified SPIONs are potential candidates to label MSCs for tracking stem cells using MRI in vivo.Acknowledgements
No acknowledgement found.References
1.Henning TD, Gawande R, Tavri S, et al. Magnetic resonance imaging of ferumoxide- labeled mesenchymal stem cells in cartilage defects: in vitro and in vivo investigations. Mol Imaging 2012; 11(3), 197–209.
2. Andreas, K.; Georgieva, R.; Ladwig, M.; Mueller, S.; Notter, M.; Sittinger, M.; Ringe, J., Highly efficient magnetic stem cell labeling with citrate-coated superparamagnetic iron oxide nanoparticles for MRI tracking. Biomaterials 2012, 33 (18), 4515-4525.
3. Sulek, S.; Mammadov, B.; Mahcicek, D. I.; Sozeri, H.; Atalar, E.; Tekinay, A. B.; Guler, M. O., Peptide functionalized superparamagnetic iron oxide nanoparticles as MRI contrast agents. J. Mater. Chem. 2011, 21 (39), 15157.
4. Preslar, A. T.; Parigi, G.; McClendon, M. T.; Sefick, S. S.; Moyer, T. J.; Haney, C. R.; Waters, E. A.; MacRenaris, K. W.; Luchinat, C.; Stupp, S. I.; Meade, T. J., Gd(III)-labeled peptide nanofibers for reporting on biomaterial localization in vivo. ACS Nano. 2014, 8 (7), 7325-7332.
Figures