2972
Intracranial vessel wall segmentation on 3D black-blood MRI using convolutional neural network1Center for Biomedical Imaging Research, Tsinghua university, Beijing, China, 2Center for Brain Disorders Research, Capital Medical University, Beijing, China, 3School of Computer Science and Technology, Beijing Institute of Technology, Beijing, China
Synopsis
Intracranial artery atherosclerosis is a major cause of stroke. manually segmenting intracranial artery vessel wall is laborious and time-consuming. we proposed an automatic intracranial artery vessel wall segmentation framework to find the centerline of the intracranial artery from SNAP images to segment the final lumen and outer-wall contours on the cross-sectional 2D slices perpendicular to the centerline.
Introduction
Intracranial artery atherosclerosis is a major cause of stroke[1]. Recently, black-blood MR imaging techniques have been proposed to better evaluate the intracranial artery atherosclerotic plaque by assessing the plaque other than luminal stenosis, such as the Volume ISotropic Turbo spin echo Acquisition (VISTA) and Simultaneous Non-contrast Angiography and intraPlaque (SNAP) techniques. However, manually segmenting intracranial artery vessel wall is laborious and time-consuming. Thus, in this study, we proposed an automatic intracranial artery vessel wall segmentation framework using a 3D convolutional neural network (CNN) to find the centerline of the intracranial artery from SNAP images and a 2D CNN to segment the final lumen and outer-wall contours on the cross-sectional 2D slices perpendicular to the centerlineMethod
Figure one shows the flowchart of the proposed framework. First, intracranial SNAP images of 100 subjects were used to automatically find the centerline. The lumen of intracranial arteries were manually segmented using Icafe[2]. Then the 3D VISTA images of these cases were automatically registered to the 3D SNAP images using FLIRT[3,4,5], and the segmentation of SNAP was mapped to VISTA images. Thus, the all the artery lumen on SNAP images has a label. In this study, labeled SNAP images of random selected 90 cases with the labeled artery lumen are used to train a 3D U-Net model [6]. And the labeled SNAP images of the left 10 cases were used as the test dataset. After the automatic lumen segmentation, the centerline of the intracranial artery can be generated automatically using a skeletonizing method[7]. Then, the 2D cross-sectional VISTA images perpendicular to the lumen centerline can be acquired using multi-planar reformation. To further segment the lumen and outer-wall in these cross-sectional images, this study further uses a 2D CNN with a manually segmented dataset. The intracranial VISTA images of 43/49 cases were used in this step. The centerline of this dataset was manually drawn, then the lumen and outer-wall in cross-sectional images were manually segmented. Similarly, labeled VISTA images of 39/42 randomly selected cases were used to train a 2D U-Net model [8]. The labeled VISTA images of the left 4 cases were used as the test dataset. Finally, the SNAP and VISTA images of one case were utilized to test the whole framework. The pixel-wise dice value ($$$2\frac{A\bigcap B}{A\bigcup B}$$$) was used to evaluate the performance of 3D lumen segmentation of 3D-CNN, the 2D lumen and outer-wall segmentation of 2D-CNN, and the final framework, with the manual segmentation as the reference.
Results
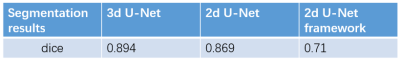
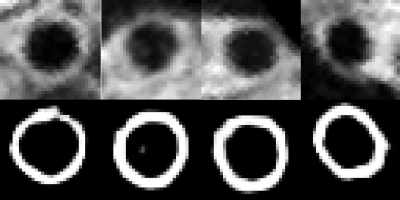
Table 1 shows the dice index of the 3D lumen segmentation of 3D-CNN, the 2D outer-wall segmentation of 2D-CNN, which were all large than 0.86. Fig. 2 shows the example of the segmentation results of the proposed method compared with the manual segmentation. A good agreement can be observed. The dice of the whole framework was 0.71 in one tested images.
Discussion and Conclusion
In this study, a framework to automatically segment the intracranial artery vessel wall using VISTA and SNAP images. The good agreement between the segmentation by the proposed automatic method and the manual segmentation suggests it could be a good tool to evaluate the intracranial atherosclerotic plaque. Although the final lumen and outer-wall were segmented purely based on 3D black-blood VISTA images, utilizing the SNAP images to find the accurate centerline of the tortuous intracranial artery is also important because SNAP has much higher lumen-wall contrast than VISTA. On the other hand, the original lumen segmentation of SNAP was not used in the final results is because SNAP tends to overestimate the lumen size. In conclusion, the proposed CNN based automatic segmentation framework has the ability to accurately segment intracranial artery vessel wall. The rough segmentation contours may be caused by the simple cut-off of the U-Net output. Further optimization of the final contours should add the active contour or level set methods on the output of U-Net.Acknowledgements
No acknowledgement found.References
1. Bogousslavsky, Julien, Guy Van Melle, and Franco Regli. "The Lausanne Stroke Registry: analysis of 1,000 consecutive patients with first stroke." Stroke 19.9 (1988): 1083-1092. 2. Chen, Li, Mahmud Mossa‐Basha, Niranjan Balu, Gador Canton, Jie Sun, Kristi Pimentel, Thomas S. Hatsukami, Jenq‐Neng Hwang, and Chun Yuan. "Development of a quantitative intracranial vascular features extraction tool on 3D MRA using semiautomated open‐curve active contour vessel tracing." Magnetic Resonance in Medicine (2017). 3. Jenkinson, M., Bannister, P., Brady, J. M. and Smith, S. M. Improved Optimisation for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage, 17(2), 825-841, 2002. 4. Jenkinson, M. and Smith, S. M. A Global Optimisation Method for Robust Affine Registration of Brain Images. Medical Image Analysis, 5(2), 143-156, 2001. 5. Greve, D.N. and Fischl, B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage, 48(1):63-72, 2009. 6. Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., & Ronneberger, O. (2016, October). 3D U-Net: learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 424-432). Springer International Publishing. 7. Ebert, D., Brunet, P., & Navazo, I. (2002). An augmented fast marching method for computing skeletons and centerlines. 8. Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 234-241). Springer, Cham.Figures
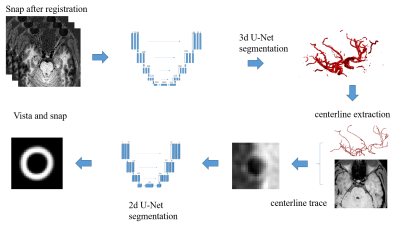
Fig1.[c1] The proposed framework for automatic intracranial artery vessel wall segmentation based on VISTA and SNAP images. The 3D U-net were used for lumen segmentation on SNAP; then the SNAP images were registered with VISTA images; the centerline of the lumen segmentation was generated and mapped to the VISTA image; the cross-sectional VISTA images perpendicular to the centerline were generated; then the 2D lumen and outer-wall segmentations were generated using a 2D U-Net model. [c1]this flow chart is not correct.