2971
Automated Segmentation of the Carotid Bifurcation using Region Growing and Support Vector Machines1Linköping University, Linköping, Sweden, 2Center for Medical Image Science and Visualization (CMIV), Linköping, Sweden, 3University Hospital Linköping, Linköping, Sweden
Synopsis
Roughly 1 in 40 deaths worldwide are caused by strokes resulting from emboli that reach the brain from ruptured atherosclerotic plaques in the carotid artery. Segmentation of the carotid artery bifurcation in MR is necessary enables further analysis. Unfortunately, this is a slow and difficult task that is often performed manually. Two segmentation methods, one based on Region Growing (RG), and one using Support Vector Machines (SVM), were implemented for segmenting the carotid bifurcation in contrast-enhanced MR Angiograms (CE-MRA). Both methods were tested quantitatively, against ground truth segmentations using the DICE and true-positive ratio (TPR) and were also scored qualitatively using visual inspection. Both methods scored highly (RG 0.890 ± 0.022, SVM 0.890 ± 0.022) using the DICE score and true-positive ratio (RG 0.938 ± 0.026, SVM 0.931 ± 0.285). During qualitative assessments, RG and SVM both scored highly with median score 4/5.
Introduction
Roughly 1 in 40 deaths worldwide are caused by strokes resulting from emboli that reach the brain from ruptured atherosclerotic plaques in the carotid artery(1). As a result, imaging the carotid bifurcation is growing in importance as we strive to predict the rupture risk of atherosclerotic plaques. Magnetic resonance imaging (MRI) permits examination of not only geometric but also compositional and hemodynamic features of carotid plaques. However, the analysis of this rich MRI data requires segmentation of the carotid artery lumen. Unfortunately, manual segmentation of the carotid arteries is a slow and difficult task with often inconsistent results. Therefore, the aim of this work was to design and test methods for automated segmentation of the carotid arteries.Method
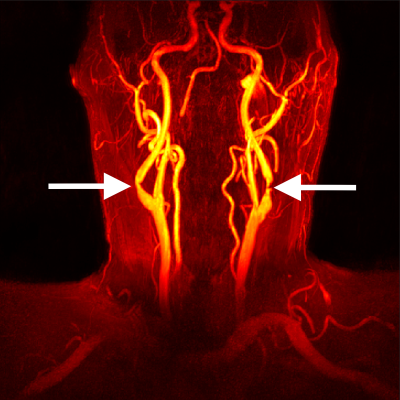
Contrast-Enhanced MR Angiograms (CE-MRA) were acquired for 25 individuals with mild or moderate stenosis as part of the SCAPIS study (scapis.se)(1). CE-MRA volumes were acquired after the injection of a gadolinium-based contrast agent, with the following scan parameters: flip angle 27°, TE = 1.8 ms, TR = 5.1 ms, in-plane spatial resolution 0.5 mm, and slice thickness 1 mm. Figure 1 shows a maximum intensity projection of the CE-MRA for a typical subject.
Two methods were implemented for the segmentation of carotid CE-MRA data: region growing (RG) and support vector machine (SVM). RG was implemented with a threshold optimization procedure that considers edge-gradients and regional intensity variance (2). The optimization finds the segmentation with minimum variance and maximum edge gradient, representing a homogeneous region with a distinct border.
A linear, hard-margin SVM segmentation method was implemented to determine whether or not voxels are within the object based on the following features: voxel intensity, mean voxel intensity in 3x3x3 neighbourhood, standard deviation in the same neighbourhood, magnitude of the gradient, magnitude of the Sobel kernel for three directions, magnitude of the Frangi filter(3) for that voxel, and the distance between that voxel and the local maximum of the Frangi filter. All features were normalized and centred around zero to remove bias. The SVM classifier used a radial basis function kernel.
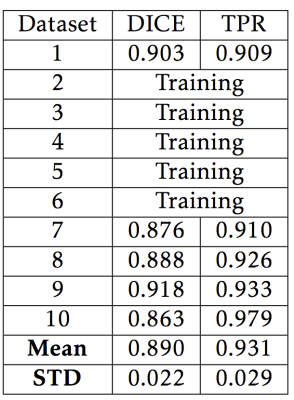
Ten datasets were manually segmented using ITK-SNAP(4) by an experienced observer, and served as ground-truth. Five of the ground-truth datasets were also used to train the SVM classifier. Segmentations were evaluated quantitatively using the DICE score and true-positive ratio (TPR). Segmentations for 15 additional subjects were qualitatively examined, and the quality of these segmentations was scored using a 5-point Likert scale by an experienced observer. Ground-truth datasets were segmented 3 times each to examine how different semi-automatically selected seed-points affected the segmentation.
Segmentations were post-processed to standardize the length of the common, internal, and external carotid arteries. Segmentations were also smoothed, and any non-contiguous regions were removed.
Results
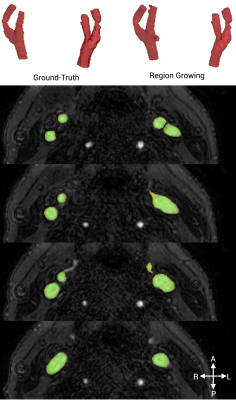
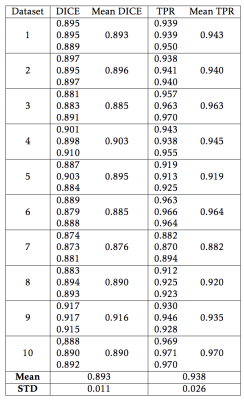
RG segmentations scored highly using both the DICE metric (mean score 0.893 ± 0.011) and TPR (mean score 0.938 ± 0.026)(Table 1). Analysis time was on the order of 2 minutes. The median qualitative score was 4/5. An example segmentation resulting from RG is shown in Figure 2.
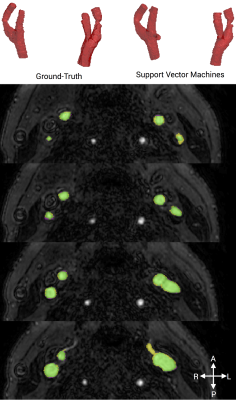
SVM segmentations similarly scored highly using both the DICE metric (mean score 0.890 ± 0.022) and TPR (mean score 0.931 ± 0.285)(Table 2). The median qualitative score was 4/5. Training, using 5 subjects, took 13 hours using a workstation with a 2.2 GHz 6 core CPU and 64 GB ram. Classifying each new dataset took approximately 10 minutes. An example segmentation from SVM can be seen in Figure 3.
Discussion
Accurate segmentations are crucial for analysis, and methods that reduce or eliminate manual input are needed for large-cohort studies. We have investigated two methods, RG and SVM, that yielded promising results. Currently, RG requires some limited user-input to begin the segmentation process, though this may be eliminated through future work. SVM segmentations could be possibly improved by increasing the number of subjects included in our training set, and through an expanded list of classification features. Future work to automatically classify segments as the common, internal, and external carotid arteries would be valuable.Acknowledgements
No acknowledgement found.References
1. Bergström G, Berglund G, Blomberg A, et al. The Swedish CArdioPulmonary BioImage Study: objectives and design. J. Intern. Med. 2015;278:645–659. doi: 10.1111/joim.12384.
2. Deng W, Xiao W, Deng H, Liu J. MRI brain tumor segmentation with region growing method based on the gradients and variances along and inside of the boundary curve. In: 2010 3rd International Conference on Biomedical Engineering and Informatics. IEEE; 2010. pp. 393–396. doi: 10.1109/BMEI.2010.5639536.
3. Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. Med. Image Comput. Comput. Assist. Interv. 1998;1496:130–137.
4. Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015.
Figures