2880
Towards a Routine Clinical Application of Chemical Exchange Sensitive MRI1Division of Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany, 3Heidelberg Institute of Radiation Oncology (HIRO), National Center for Radiation Research in Oncology (NCRO), Heidelberg, Germany, 4Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 5Center for Biomedical Imaging, NYU Langone Medical Center, New York, NY, United States, 6Division of X-Ray Imaging and Computed Tomography, German Cancer Research Center (DKFZ), Heidelberg, Germany, 7Max Planck Institute for Biophysical Chemistry, Goettingen, Germany
Synopsis
Chemical exchange saturation transfer (CEST) and chemical exchange sensitive spinlock (CESL) were shown to have potential to provide molecular information for diagnosing a wide range of diseases. However, the lack of standardized acquisition protocols and freely available post-processing software prohibited the widespread application of these promising techniques until now. In this work, we present a modularly designed CEST/CESL preparation block that is easy to operate and can be used with arbitrary MRI readouts. Further, we developed and provide a C++ based open-source software that offers many CEST/CESL specific functionalities for the post-processing of the acquired data.
Introduction
Chemical exchange (CE) sensitive MRI techniques like chemical exchange saturation transfer (CEST) or chemical exchange sensitive spinlock (CESL) enable imaging of low concentrated compounds with spatial resolutions typical for conventional water imaging. This makes CE sensitive MRI a promising technique for diagnosis of cancer or neurodegenerative diseases.1 However, due to the time-consuming implementation of the MRI sequence and the complex analysis processes, CE sensitive MRI did not yet made its way into clinical routine. Thus our aim was to develop a CEST/CESL sequence that is easy to implement and operate, and to provide open-source software for the post-processing of the acquired data.Theory
CE sensitive MRI sequences are composed of a magnetization preparation and a data acquisition part. The simplest RF preparation in CEST-MRI is a rectangular pulse that is applied for several seconds. However, safety issues and technical limitations of whole-body MR scanners require the use of more advanced preparation approaches consisting of several shorter pulses of more sophisticated shapes.2 The development of such advanced preparation blocks is quite complex and time-consuming.
The asymmetry analysis (MTRasym)3 is the simplest and most commonly utilized CEST metric. Yet, it already requires the normalization and correction for B0-inhomogeneities of the data. More sophisticated CEST metrics like the apparent exchange-dependent relaxation (AREX)4 and/or additional correction algorithms (e.g. for spatial variations of B1) further require the acquisition of B1 and/or T1 maps.5
Methods
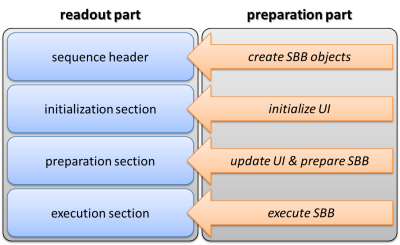
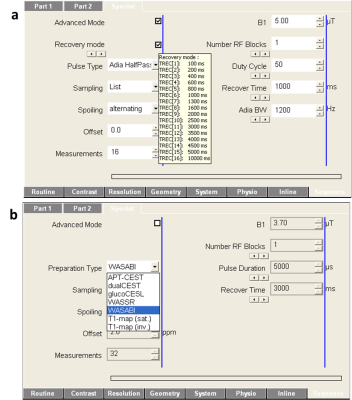
The pulse sequence was developed within the Integrated Development Environment for MR Applications (IDEA) of Siemens (Siemens Healthcare, Germany). The preparation part was designed in a modular manner and consists of a sequence building block (SBB) and a user interface (UI). The following parameters can be adjusted in the UI: number, duration, distance, power and shape of the saturation pulses, number of offsets and sampling type (e.g. ′regular distance′, ′read txt-file′), recovery times, spoiling options and the parameters of the adiabatic pulses, which are used for spinlock6 and T1 measurements. Due to its modular design, the preparation part can be merged with almost any readout sequence with just little modifications (Fig. 1).
Processing of the resulting images was done with the Medical Imaging Interaction Toolkit (MITK)7. MITK is a C++ based library and application framework. It provides a suite of tools for many common image processing needs such as segmentation, image registration, fitting functionality and read/write support for a wide variety of image formats. It also provides specialized functionality for specific MR applications such as diffusion-weighted MRI8. It was extended to support the presented CEST/CESL sequence and provide CEST/CESL specific features.
Results
The developed preparation block was successfully merged with various readout sequences. These were checked for functionality on several Siemens MR-systems operated under software versions between IDEA VB17 and VE11. The sequences allow the acquisition of arbitrary CEST/CESL contrasts and all relevant parameter maps. This includes the simultaneous acquisition of B0 and B1 maps by means of WASABI9 and the acquisition of T1 maps using adiabatic inversion or saturation recovery. Further, the implemented basic-mode allows switching between these and other predefined preparation-types easily (Fig. 2).
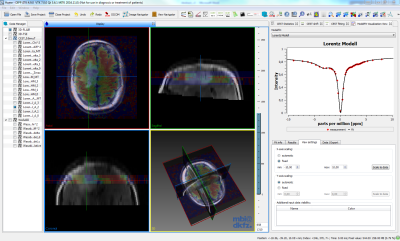
The developed analysis and visualization software MITK-CEST extends the general MITK workbench with several CEST/CESL features and a specific UI that was designed in cooperation with radiologists. MITK-CEST enables the intuitive visualization and interactive exploration of frequency- (e.g. CEST, WASABI) or time-dependent (e.g. T1, dynamic glucose-enhanced CEST/CESL) data and derived information (Fig. 3). Other CEST/CESL functionalities are amongst others normalization, B0-correction, fitting (e.g. T1, WASABI, Multi-Lorentzian) and the calculation and visualization of different contrasts like MTRasym or AREX.
Discussion
The CEST/CESL SBB was developed for Siemens platforms, but a similar modular design should be feasible for other platforms as well. The developed SBB allows acquiring the CEST/CESL contrasts and B0, B1 and T1 maps with the same sequence. The basic-mode allows even less experienced users to operate the sequence. Up to now, all freely available CEST tools10,11 are implemented in Matlab (MathWorks, USA) and thus require a Matlab license. Moreover, in contrast to MITK-CEST, these tools are not optimized for clinicians. The integration of CEST/CESL functionalities in MITK is also beneficial, because the well-established image processing tools like segmentation, ROI-analysis or motion-correction can be reused. The support for multiple imaging modalities further enables the co-registration and comparison with CT or PET data. Providing the CEST/CESL SBB and the MITK-CEST software will make it easier for researches to start with CEST/CESL MRI and thus may lead to a more widespread application. Further, it leads to standardized acquisition and evaluation protocols and thus to data that can be compared more easily.Acknowledgements
No acknowledgement found.References
1. Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J. Magn. Reson. Imaging 2017:1–16.
2. Sun PZ, Wang E, Cheung JS, Zhang X, Benner T, Sorensen AG. Simulation and optimization of pulsed radio frequency irradiation scheme for chemical exchange saturation transfer (CEST) MRI-demonstration of pH-weighted pulsed-amide proton CEST MRI in an animal model of acute cerebral ischemia. Magn. Reson. Med. 2011;66:1042–8.
3. Zhou J, Payen J-F, Wilson D a, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat. Med. 2003;9:1085–90.
4. Zaiss M, Xu J, Goerke S, Khan IS, Singer RJ, Gore JC, Gochberg DF, Bachert P. Inverse Z-spectrum analysis for spillover-, MT-, and T1 -corrected steady-state pulsed CEST-MRI--application to pH-weighted MRI of acute stroke. NMR Biomed. 2014;27:240–52.
5. Windschuh J, Zaiss M, Meissner J-E, Paech D, Radbruch A, Ladd ME, Bachert P. Correction of B1-inhomogeneities for relaxation-compensated CEST imaging at 7 T. NMR Biomed. 2015;28:529–37.
6. Schuenke P, Koehler C, Korzowski A, Windschuh J, Bachert P, Ladd ME, Mundiyanapurath S, Paech D, Bickelhaupt S, Bonekamp D, Schlemmer H-P, Radbruch A, Zaiss M. Adiabatically prepared spin-lock approach for T1ρ-based dynamic glucose enhanced MRI at ultrahigh fields. Magn. Reson. Med. 2016;78:215–225.
7. Nolden, M., Zelzer, S., Seitel, A., Wald, D., Müller, M., Franz, A. M., Maleike, D., Fangerau, M., Baumhauer, M., Maier-Hein, L., Maier-Hein, K. H., Meinzer, H.-P. & Wolf, I. The Medical Imaging Interaction Toolkit: challenges and advances : 10 years of open-source development. Int. J. Comput. Assist. Radiol. Surg. 8, 607–20 (2013).
8. Fritzsche, K. H., Neher, P. F., Reicht, I., van Bruggen, T.,
Goch, C., Reisert, M., Nolden, M., Zelzer, S., Meinzer, H. P. & Stieltjes,
B. MITK diffusion imaging. Methods Inf. Med. 51, 441–448 (2012).
9. Schuenke P, Windschuh J, Roeloffs V, Ladd ME, Bachert P, Zaiss M. Simultaneous mapping of water shift and B1 (WASABI) - Application to field-Inhomogeneity correction of CEST MRI data. Magn. Reson. Med. 2016;77:571–580.
10. Max Planck Institute for Biological Cybernetics & German Cancer Research Center. Cest Sources. http://cest-sources.org. Accessed November 8, 2017.
11. Kennedy Krieger Institute. CEST Data Processing Tools.http://godzilla.kennedykrieger.org/CEST. Accessed November 7, 2017.
12. Goerke S, Zaiss M, Windschuh J, Klika KD, Bachert P. Two dimensional CEST spectroscopy of proteins. Proc. Symp. Chem. Exch. Satur. Transf. PENN-CEST 2015.
13. Schuenke P, Paech D, Koehler C, Windschuh J, Bachert P, Ladd ME, Schlemmer H-P, Radbruch A, Zaiss M. Fast and Quantitative T1ρ-weighted Dynamic Glucose Enhanced MRI. Sci. Rep. 2017;7:42093.
14. Kim M, Gillen J, Landman B a, Zhou J, van Zijl PCM. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med. 2009;61:1441–50.
Figures