2877
Sulcal ridge alignment for laminar fMRI at 7T1Spinoza Centre for Neuroimaging, Amsterdam, Netherlands, 2Netherlands Institute for Neuroscience, Amsterdam, Netherlands, 3Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 4Psychology, University of Amsterdam, Amsterdam, Netherlands
Synopsis
Laminar fMRI at 7T typically involves imaging small slabs of cortex, and requires precise alignment of the anatomical and functional data to transfer intra-cortical depth information to the fMRI data. We present a method taking advantage of the high resolution of the fMRI data and extracted sulcal patterns.
Introduction
Recent advances in high field functional MRI have made it possible to zoom in below the millimeter, which opens the possibility to probe functional differences along the depth of the cortex1,2. However, doing this accurately requires one to align anatomical and functional data with utmost precision, to reduce the variance and more importantly avoid systematic biases when attributing voxels to a given cortical depth. While strategies based on unwarping of the fMRI signal and using additional image slabs of the anatomy can be successful, they require additional imaging and complex post-processing. We propose here a simpler method based on the detection of sulcal patterns directly from the fMRI data to be matched with the same patterns extracted from anatomical MRI.Methods
Image acquisition: We collected structural and function MRI in five subjects on a Phillips 7 Tesla scanner with the following sequences: a 3D EPI at 0.8 mm isotropic resolution centered on the parietal lobe (FOV 150 * 169 * 24, TR/TE = 54/28 ms, EPI factor = 27, SENSE(AP) = 4, slice oversampling volume 1.28, volume acquisition time 4.1s.) and a whole brain MP2RAGE image at 0.64 mm isotropic resolution (TI1/2 0.8/2.7s, FA1/2 7/5 deg, TE 62ms, TR 5.5s).
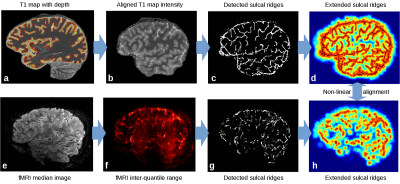
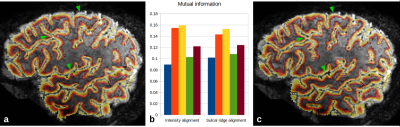
Processing: Anatomical quantitative T1 images were estimated from the MP2RAGE3. Non-brain regions were masked with an intensity-based skull stripping algorithm designed for 7T, and the images were further processed to obtain cortical surfaces and cortical depth estimates4,5 (Fig.1a). After coarse alignment based on scanner coordinates (Fig 1b), sulcal CSF was then extracted as 2D ridges on the skull-stripped T1 maps with a recursive ridge filter (RRF) originally designed for extracting vasculature6. In this version, the ridge filter goes through a single loop in order to detect 2D structures, followed by the same diffusion process applied to a 3-voxel neighborhood of the ridge center point (Fig. 1c). Functional images were motion-corrected in SPM (http://www.fil.ion.ucl.ac.uk/spm/), and the temporal median and inter-quartile range (IQR) were extracted. The brain region was estimated by a 2D slab version of the previous skull stripping algorithm applied to the median (Fig. 1e). The IQR exhibits generally high values inside the CSF and cortical veins due to CSF pulsation and venous BOLD fluctuations (Fig. 1f). It was used to detect 2D ridges as for the T1 map above (Fig. 1g), and sulcal ridge structures from both images were smoothed over neighboring voxels to a maximum of 8 mm (Fig.1d,h). The smoothed images were co-registered non-linearly with the SyN algorithm of ANTs7 after initialization using the scanner-provided orientation information. In order to test the quality of the fit, the sulcal ridge registration method was compared to the registration of the skull-stripped median fMRI and quantitative T1 map with ANTs. Results were quantified by measuring mutual information restricted to a region including the cortex and dilated by two voxels to include the WM and CSF boundaries (Fig.2b).
Results and Discussion
While the alignment quality was globally similar between both techniques, the sulcal ridge alignment improved the result in the lowest mutual information cases. Qualitatively speaking, the ridge alignment improved matching of the CSF boundary in many regions, increasing the visibility of vessels in sulci and the alignment of gyri at the outer surface of the brain (Fig. 2a,c). In regions with stronger distortions such as the frontal lobes, neither method was able to fully resolve the difference, indicating that initializing the alignment with an fMRI distortion correction might still be beneficial.Conclusion
The sulcal ridge pattern is complex and highly tied to the cortical geometry, thus it provides precise guidance to match cortices independently of geometric distortions. Our proposed alignment technique harnesses this pattern to align cortical boundaries from the fMRI and anatomy precisely, a particularly critical step for laminar fMRI. Further improvements may be reached with a more refined detection of fMRI sulcal patterns, for instance combining information from the fMRI median and IQR in the filtering, or from a distortion-matched EPI-based T1 map. The overall approach is general and extensible, for instance to matching accurately repeated scans from ultra-high-resolution imaging experiments.Acknowledgements
We would like to thank Natalia Petridou, Alessio Fracasso and Kerrin Pine for stimulating discussions on the question of high-precision alignment. The research was funded by an ERC-StG grant VicariousBrain 312511 to CK and a grant from the BIAL foundation to VG and RB.References
1. van der Zwaag, W., Schäfer, A., Marques, J.P., Turner, R., Trampel, R., 2016. Recent applications of UHF-MRI in the study of human brain function and structure: a review: UHF MRI: Applications to Human Brain Function and Structure. NMR in Biomedicine 29, 1274–1288. doi:10.1002/nbm.3275
2. Dumoulin, S.O., Fracasso, A., van der Zwaag, W., Siero, J.C.W., Petridou, N., 2017. Ultra-high field MRI: Advancing systems neuroscience towards mesoscopic human brain function. NeuroImage. doi:10.1016/j.neuroimage.2017.01.028
3. Marques, J.P., Kober, T., Krueger, G., Zwaag, W. van der, Moortele, P.-F.V. de, Gruetter, R., 2010. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 49, 1271–1281. doi:DOI: 10.1016/j.neuroimage.2009.10.002
4. Bazin, P.-L., Weiss, M., Dinse, J., Schä̈fer, A., Trampel, R., Turner, R., 2014. A computational framework for ultra-high resolution cortical segmentation at 7 Tesla. NeuroImage 93, 201–9.
5. Waehnert, M.D., Dinse, J., Schäfer, A., Geyer, S., Bazin, P.-L., Turner, R., Tardif, C.L., 2016. A subject-specific framework for in vivo myeloarchitectonic analysis using high resolution quantitative MRI. NeuroImage 125, 94–107. doi:10.1016/j.neuroimage.2015.10.001
6. Bazin, P.-L., Plessis, V., Fan, A.P., Villringer, A., Gauthier, C.J., 2016. Vessel segmentation from quantitative susceptibility maps for local oxygenation venography. IEEE, pp. 1135–1138. doi:10.1109/ISBI.2016.7493466
7. Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C., 2008. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis 12, 26–41.
Figures