2865
High Resolution Restoration of Neonatal Images: Matlab Based Framework1Department of Radiology and Medical Informatics, University of Geneva, Genève, Switzerland, 2School of Engineering, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 3Department of Pediatrics, University Hospitals of Geneva, Genève, Switzerland, 4Biomedical Imaging Group, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Synopsis
A MATLAB based graphical user interface (GUI) was created to apply super resolution (SR) technique on low resolution neonate MR images for obtaining high resolution volume. The user has options to compute HR volume with registration and different reconstruction and regularization methods. In quantitative analysis section, root mean square error, signal-to-noise ratio values could be computed.
Purpose/Introduction
Super resolution reconstruction (SR) is a technique that combines set of low resolution (LR) images and produce higher resolution (HR) images [1,2]. Due to time limitations and motion sensitivity in neonatal MRI, SR method could be an alternative to lengthy high-resolution acquisitions. Several strategies to reduce scan time can be considered, such as acquiring thick slices, parallel or orthogonal stacks or in-plane low resolution images. These multiple low-resolution MRI stacks can be combined, after applying registration and reconstruction algorithm to obtain an HR volume.
In order to cope with various acquisition strategies, different reconstruction methods and regularization options, we propose a MATLAB (The MathWorks, Inc.) based graphical user interface (GUI) to automatize neonatal image restoration to obtain motion-free high-resolution volume.
Method
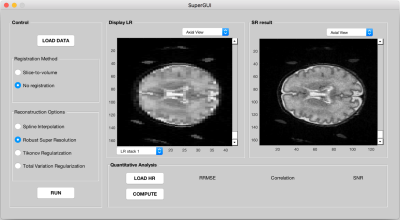
Figure 1 shows the MATLAB GUI for super resolution reconstruction techniques. This interface is composed of three parts: registration, restoration and analysis. As an input, either orthogonal LR scans or through slice shifted low resolution stacks can be used.
Depending on acquisition scheme, user selects appropriate registration method. In the case of through slice shifted LR stacks, it does not need to be registered if the motion is negligible. In the case of orthogonal LR stacks the registration will be performed with slice-to-volume registration method [3,4]. Image Restoration process comes with four different options: Spline interpolation, Robust Super resolution [5], super resolution with regularization, either with Tikhonov [6,7] or Total variation [8,9]. The quantitative analysis is optional when a reference HR image is available.The user can compute relative root mean square error (RRMSE), linear correlation and signal to noise (SNR) values between SR result and ground truth images.
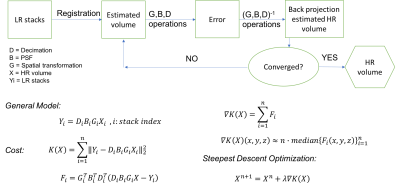
Figure 2 shows the general diagram of SR method. The algorithm minimises the error while reconstructing the HR volume from the limited number of LR stacks with gradient based iterative optimization. Point spread function (PSF) is adjusted depending on the acquisition scheme (slice profile or Gaussian).
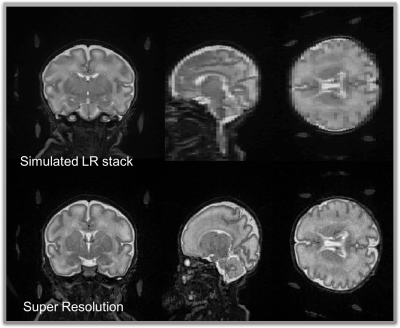
This GUI was used on two different neonate MRI dataset. First on simulated LR neonate dataset computed from HR: HR T2w coronal images acquired in 3T MRI scanner (Prisma-Siemens, Erlangen, Germany) using 64 channel receiver head coil. High resolution slices consisted of Turbo spin echo (TSE) multiband sequence with TR = 5100ms , TE = 153 ms, in plane resolution 0.8 mm and slice thickness 1 mm, acquisition time was 2 min 22 sec. We consider high-resolution images as ground truth images. Three orthogonal stacks (coronal, sagittal, axial) with 0.8 mm in plane and 3 mm slice thickness were simulated from the ground truth image by applying proper homogenous 6 degree of freedom reslicing matrices, PSF and downsampling operations. Second, we tested the interface on a clinical neonate dataset: four through-plane low resolution T2w TSE images were acquired in 3T MRI scanner (Prisma-Siemens, Erlangen, Germany) with parameters of TR = 5100 ms , TE = 153 ms, acquisition time for each stacks was 41 sec, in plane resolution 0.8 mm and slice thickness 3 mm with 0.65 mm through-plane shift between each image stacks.
Results
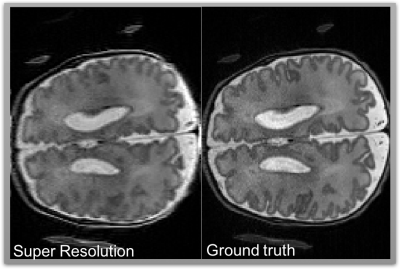
Figures 3 and 4 show the SR results from two different datasets, simulated and clinical, respectively. In both cases SR gives visually and quantitatively good results and that are very close to gold standard HR volumes. In the GUI there are two figure boxes. While left shows low resolution images, right displays final SR images after user runs the process. Both LR and HR volumes can be seen in three different orientations (coronal, sagittal, axial planes). This helps user to navigate and review the different stacks and disregard unnecessary stacks.Discussion and Conclusion
The presented MATLAB based GUI provides flexible, fast, interactive and user-friendly platform to display, register and reconstruct low resolution MR images. The user has option to check input data and result, visually. So far quality of images is decided visually, in the future we plan to incorporate an automatic assessment procedure.
The designed GUI is a good and quick option when there is quantitatively high amount of subjects to analyse. It is based on DICOM standard that allows reconstructed images to be incorporated in the clinical database.
Acknowledgements
We would like to acknowledge to Dr. Joana Sa De Almeida for recruitment of the babies.References
[1] M. Elad, A. Feuer. 1997. Restoration of single super-resolution image from several blurred, noisy and down-sampled measured images IEEE Trans Image Process 6:1646–1658.
[2] M.Irani and S.Peleg, “Improving resolution by image registration,” CVGIP: Graph. Models Image Process (1991), vol. 53, pp. 231–239.
[3] F. Rousseau, O. A. Glenn, B. Iordanova, C. Rodriguez-Carranza, D. B. Vigneron, J. A. Barkovich, C. Studholme, Registration-based approach for reconstruction of high- resolution in utero fetal MR brain images, Academic Radiology 13 (9) (2006) 1072–1081.
[4] A. Gholipour, J. Estroff, S. Warfield, Robust super-resolution volume reconstruction from slice acquisitions: Application to fetal brain MRI, Medical Imaging, IEEE Transactions on 29 (10) (2010) 1739–1758.
[5] A. Zomet, A. Rav-Acha, S. Peleg. 2001. Robust super resolution, In: Proc. Int Conf Computer Vision and Pattern Recognition (CVPR), Dec 2001, Vol. 1, pp. 645– 650.
[6] A. N. Tikhonov and V. A. Arsenin. Solution of ill-posed problems. Winston & Sons, Washington, 1997.
[7] A.Gholipour, S.K. Warfield, Super-resolution reconstruction of fetal brain MRI, in: MICCAI Workshop on Image Analysis for the Developing Brain (IADB’2009), 2009, pp. 45–52
[8] L. Rudin, S. Osher, E. Fatemi. 1992. Nonlinear total variation based noise removal algorithms. Physica D 60:259–268.
[9] S. Farsiu, D. Robinson, M. Elad, P. Milanfar. 2003a. Fast and robust Super-Resolution. Proc 2003 IEEE Int Conf on Image Process 2003, pp. 291–294.
Figures