2844
A High Performance Computing Cluster Implementation Of Compressed Sensing Reconstruction For MR Histology1Center for In Vivo Microscopy, Department of Radiology, Duke University, Durham, NC, United States, 2GE Healthcare, Salt Lake City, UT, United States
Synopsis
We report the generation of a software pipeline for accelerated MR image reconstruction in a high-performance computing environment, motivated by the shift in time demands from the acquisition to the computational burden of reconstruction in compressed sensing.
Introduction
In the world of Magnetic Resonance Histology1 (MRH), Diffusion Tensor Imaging (DTI) has opened the door to new quantitative methods such as Voxel-Based Analysis (VBA) and connectomics. Unfortunately, already lengthy acquisition times are exacerbated when sampling 45+ diffusion angles, requiring >4 days2. Even with fixed post mortem specimens, such times are not practical for routine use. Recently, MRH Compressed Sensing (CS) acquisition protocols were used to achieve acceleration factors (AF) of 4-16x, thus enabling a throughput of multiple specimens per day3. This shifts the rate-limiting step to the computationally demanding iterative CS image reconstruction (CSR) process. However, MR Histology poses a distinct challenge here: for the relatively small (by MRH standards) isotropic image array size of 512x256x256 with double-precision complex data, the working footprint of a 51 volume DTI scan is over 25 GB. Even worse, it takes ~12 seconds to reconstruct each of the 26112 slices—about half a week’s worth of computation—negating any gains realized by the accelerated acquisition. We present here a CSR pipeline deployed in a high-performance computing (HPC) cluster. By taking advantage of the parallel nature of slice-wise CSR, the total reconstruction time can be reduced 60-150x and meet the demands of CS-MRH on a routine basis.Methods
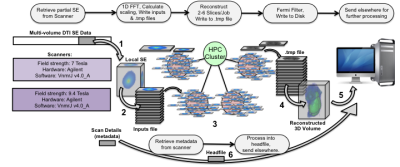
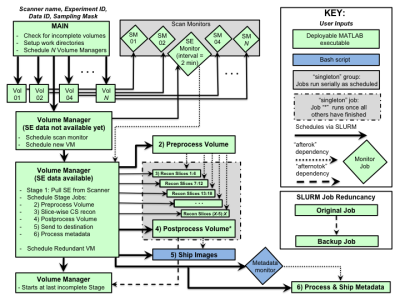
In Figure 1, Spin-echo (SE) data passes through 6 stages to become a fully reconstructed 4-5D image, ready for further processing streams such as tensor estimation and SAMBA, a VBA and connectomics pipeline for small animals4. Written in MATLAB and Linux bash, the code uses SLURM5 for dispatching jobs to the cluster, with SLURM job dependencies orchestrating their execution. MATLAB code for reconstructing CS MR images via regularization of an objective function based on wavelet transforms and total variation6,7 was parsed into minimal units of work and compiled as executables to avoid cluster-wide licensing limits. Custom MATLAB code interfaces with the scanners to facilitate streaming and validation during the long scans. Figure 2 outlines the workflow. Each diffusion image has its own Volume Manager, which executes Stage 1 directly and schedules the other 5. The addition of small data monitoring jobs enables the efficient streaming mode where CSR for a volume begins as soon as that portion of the scan has been acquired. Scheduling backups for each job, with special handling for the Volume Managers, provides an additional layer of robustness.Results
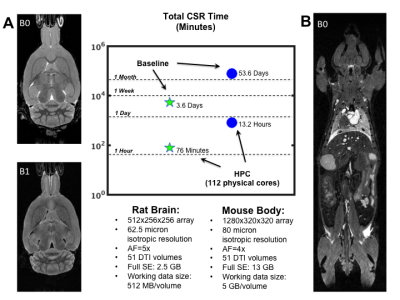
The value of the pipeline is demonstrated in Figure 3 via two CS-DTI data sets: a rat brain (A) and a whole mouse body (B). As a baseline for comparison, original reconstruction times are given for unmodified code running on a single node (single-threaded by default). The HPC times are running on 112 of 176 physical cores spread across a cluster consisting of 10 nodes with 256 GB RAM for each node. The fractional usage of cores represents a steady load on the cluster without completely monopolizing resources.
In the rat brain CSR was accelerated by 68x, while a factor of 98x was observed for the whole mouse body when utilizing 112 cores. If necessary, this can be pushed to ~150x by using the entire cluster, as it is directly proportional to the number of cores. Reductions in the pre- and post-processing times (not shown) were also realized via code optimization, leading to 2-4x speedup for those stages. Importantly, the order of 1 and 13 hours are acceptable times for the respective scans, without requiring the cluster’s full resources.
Discussion
The HPC implementation of CSR is vital to producing images on the same order as the accelerated acquisition time. In addition to higher throughput and angular resolution, streaming CSR allows for quality assurance in a relevant timeframe and rapid prototyping of novel DTI protocols. For example, it has recently been used in a multi-shell sampling scheme with over 400 diffusion measurements8. Two potential ways to further reduce reconstruction times is to move from a fixed number of iterations to a convergence model, and GPU cluster implementation, both of which are left to future work. The code is publicly available via GitHub9.Conclusion
We have developed a cluster-based compressed sensing reconstruction pipeline specifically to handle the challenges presented by 4-5 dimensional MR Histology studies. We have demonstrated achievable speedups of 60-100x for brain and whole body imaging of rodents. Reconstruction in a reasonable time is possible even when few cluster nodes are available. This application is relevant for high dimensional image arrays, and/or multi-volume acquisitions as is the case for multi-echo and diffusion imaging protocols.Acknowledgements
All work was performed at the Center for In Vivo Microscopy, supported by NIH awards: Office of the Director 1S10ODO10683-01, NIH/NINDS 1R01NS096720-01A1 (G Allan Johnson). We also gratefully acknowledge NIH support for our research through K01 AG041211 (Badea).References
1. Johnson GA, Benveniste H, Black R, Hedlund L, Maronpot R, Smith B. Histology by magnetic resonance microscopy. Magnetic resonance quarterly (1993). Vol. 9, pp 1-30.
2. Calabrese E, Badea A, Cofer G, Qi Y, Johnson GA. A Diffusion MRI Tractography Connectome of the Mouse Brain and Comparison with Neuronal Tracer Data. Cerebral Cortex (2015), Vol. 25, Issue 11, pp 4628–4637.
3. Wang N, Cofer G, Anderson RJ, Dibb R, Qi Y, Badea A, Johnson GA. Compressed Sensing to Accelerate Connectomic Histology in the Mouse Brain, In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, Hawaii, USA, April 2017 (Abstract 1778).
4. SAMBA: a Small Animal Multivariate Brain Analysis pipeline, code available at: https://github.com/andersonion/SAMBA/.
5. Yoo AB, Jette MA, Grondona M. 2003. Slurm: Simple linux utility for resource management. Job Scheduling Strategies for Parallel Processing Job Scheduling Strategies for Parallel Processing. JSSPP 2003. Lecture Notes in Computer Science, Vol. 2862, pp 44-60.
6. Lustig M, Donoho D, Santos J, Pauly J. Compressed Sensing MRI. IEEE Signal Processing Magazine (2008), Vol. 25, Issue 2, pp 72 – 82.
7. Berkley Advanced Reconstruction Toolbox (BART), open source code available at https://mrirecon.github.io/bart/.
8. Wang N, Zhang J, Anderson RJ, Cofer G, Qi Y, Johnson GA. High-resolution Neurite orientation dispersion and density imaging of mouse brain using compressed sensing at 9.4T. Abstract submitted to the 26th Annual Meeting of ISMRM, Paris, France, June 2018.
9. Code available at: https://github.com/andersonion/MR_compressed_sensing_HPC_recon/
Figures