2840
Random Forest based Calf Muscle Segmentation from MR data incorporating Prior Information1Section on Experimental Radiology, Department of Diagnostic and Interventional Radiology, University Hospital of Tübingen, Tübingen, Germany, 2Institute of Signal Processing and System Theory, University of Stuttgart, Stuttgart, Germany
Synopsis
Delineation of muscle structures from MR images is an intricate but essential step for quantitative morphological assessment in many areas. In this work segmentation of muscles in the right calf from 2D MR data has been performed. Since challenging conditions prevail, prior information was incorporated in a Machine Learning driven approach. Versatile Random Forests were employed making use of annotated atlases as well as defined landmarks. It was demonstrated that incorporation of this prior information results in a feasible and fully automatic muscle segmentation.
Introduction
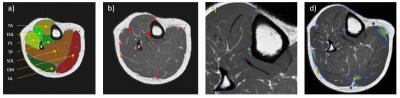
Reliable delineation of muscle structures from MR datasets is a fundamental prerequisite for quantitative assessment of morphological markers in many areas, including determination of muscle volume for disease progression and correlation analysis in metabolic diseases. However, manual/semi-automatic segmentation of muscles is cumbersome and expert-dependent. Thus, segmentation algorithms based on few annotated data which facilitate automatic analysis of intricate muscle structures are of interest. In this work, 2D MR images of the right calf of healthy volunteers are considered. In this setting, a suitable algorithm has to account for diverse factors such as vast variations in shape and relative position of each muscle, but also for the lack of distinctive texture or intensity features, rendering common schemes infeasible. We propose a Random Forest1 (RF) driven two-stage approach that incorporates beneficial aspects by means of annotated atlases (Figure 1a) and prominent predefined landmarks (Figure 1b) to gain necessary prior information.Methods
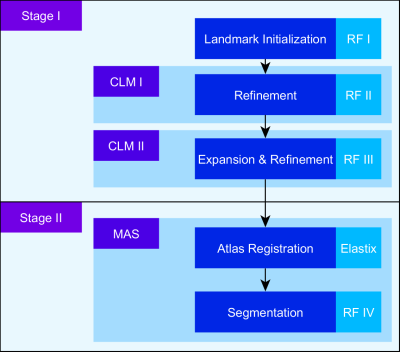
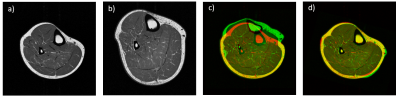
The proposed pipeline depicted in Figure 2 consists of two distinct stages. In the first stage, prominent landmarks are identified. This allows for a guided registration of an atlas set in the second stage. Thus, meaningful spatial correspondence is established, allowing subsequent segmentation. Both stages are consecutively driven by use of RFs, an efficient Machine Learning (ML) algorithm for a variety of applications. The employed RF architecture2,3 performs landmark identification (RF I), positional refinement of found landmarks (RF II/III, see Figure 1c,d) and segmentation into muscles (RF IV) based on given features. We make use of features extracted from Gabor filters and intensity features from the surrounding neighborhood to capture sufficient texture and context information. To alleviate the initialization of prominent points, RF I learns to identify a subset of all employed landmarks which is subsequently expanded to the full set in RF III. To capture spatial relationships between the landmarks, RF II & III are embedded in a statistical model, called Constrained Local Model (CLM).4 Similar to shape models,5 CLMs restrict identified positions to plausible solutions. RF IV allows for the encoding of prior information by the registered atlas labels, which is known as learning based Multi-Atlas Segmentation (MAS).6 Additional features of the atlases are created to take annotations and relative spatial positions into account. Atlas registration is performed by the Elastix toolbox,7 which provides rigid, affine and non-rigid registration algorithms without (unguided) and with (guided) incorporation of predefined points. The importance of guidance is visualized in Figure 3.Results & Discussion
45 anatomical images have been acquired on a 1.5 T (Magnetom Sonata, Siemens Healthcare GmbH, Erlangen, Germany) via an extremity coil with a 2D transverse TSE sequence (FOV: 180x180 mm², 512x512 pixel (reconstructed), slice-thickness: 6 mm, TE: 16 ms, TR: 650 ms, ETL: 3, PF: 0.8, BW: 250 Hz/px). In Figure 1a the seven different muscle (groups) considered for segmentation are shown (m. gastrocnemius medialis (GM), gastrocnemius lateralis (GL), soleus with flexor digitorum longus (SOL), tibialis anterior (TA), tibialis posterior (TP), extensor digitorum/hallucis longus (EL) and peroneus longus (PL). Alongside, the predefined landmarks are indicated in Figure 1b. For testing a leave-5-out cross-validation was performed. We considered only 5 patients of the respective training set to demonstrate that our learning based method is feasible with few annotated data. Besides the learning-based approach of stage II a traditional MAS with majority voting (MV) was employed as baseline. In addition the effect of unguided and guided atlas registration was investigated. We compared the results of manually defined landmarks (guided) with landmarks identified by stage I (voted). Resulting segmentations are illustrated in Figure 4. Correlation with the manual segmentation is especially given for guided variants, which is reflected in high Dice Similarity Coefficients (DSC) and low Average Surface Distances (ASD) in Figure 5. The landmarks were reliably found by stage I, but a misalignment of 20.34 ± 10.78 mm was present. Despite the high variability present in the training set, promising results could be achieved. However, establishing atlas correspondence remains difficult for some regions even with provided landmarks as seen by poor values for EDL.Conclusion
A RF driven segmentation pipeline has been proposed,
demonstrating the feasibility of automatic calf muscle segmentation under
challenging conditions. The scheme could be refined by incorporating an atlas
matching scheme,8 alleviating the segmentation by providing better
quality of atlas features. Also more landmarks could be considered to further
enhance atlas alignment. Further investigations incorporating more annotated
data along with atlas selection schemes has to be performed.Acknowledgements
No acknowledgement found.References
1. Criminisi, Antonio, Jamie Shotton, and Ender Konukoglu. "Decision forests: A unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning." Foundations and Trends® in Computer Graphics and Vision 7.2–3 (2012): 81-227.
2. Glocker, Ben, et al. "Joint classification-regression forests for spatially structured multi-object segmentation." Computer Vision–ECCV 2012 (2012): 870-881.
3. Lassner, Christoph, and Rainer Lienhart. "The fertilized forests Decision Forest library." Proceedings of the 23rd ACM International Conference on Multimedia. ACM, (2015):681-684.
4. Cootes, Tim F., et al. "Robust and accurate shape model fitting using random forest regression voting." European Conference on Computer Vision. Springer Berlin Heidelberg, (2012):278-291.
5. Heimann, Tobias, and Hans-Peter Meinzer. "Statistical shape models for 3D medical image segmentation: a review." Medical Image Analysis 13.4 (2009): 543-563.
6. Lee, Hoileong, et al. "Learning-Based Multi-atlas Segmentation of the Lungs and Lobes in Proton MR Images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, (2017):108-115.
7. Klein, Stefan, et al. "Elastix: a toolbox for intensity-based medical image registration." IEEE Transactions on Medical Imaging 29.1 (2010): 196-205.
8. Giraud, Rémi, et al. "An optimized patchmatch for multi-scale and multi-feature label fusion." NeuroImage 124 (2016): 770-782.
Figures