2806
Deep-learned STIR imaging via Deep Learning with multi-contrast MRI1Yonsei University, Seoul, Republic of Korea, 2Philips Korea, Seoul, Republic of Korea
Synopsis
The goal of this study is to make STIR MRI using deep learning with multi-contrast MRI. First, we simulated the phantom image created by the bloch equation, which is the basic formula for making MRI, and confirmed that the convolution neural network learns the bloch equation. We also showed the feasibility of making STIR image with in-vivo T1- and T2-weighted, and GRE images in the knee.
Purpose
MRI has various contrasts depends on the purpose of diagnosis.(e.g. T1-, T2- and PD-weighted images, GRE, FLAIR, STIR) In general, four or five images are obtained once on MRI scan, and acquisition time differs from each sequences. Since it takes a long time to acquire all of images, if we can make the one image from the rest, the acquisition time can be drastically reduced. Therefore, we proposed a method of making a new contrast MRI with multi-contrast MRI1,2,3.
Introduction
Among the various kinds of MRI, STIR image is often used in the knee. STIR image is a kind of inversion recovery equation, which suppresses the fat signal and increases the contrast of tumor or inflammatory tissue around the fat. In addition, since fat and bone are similar to T1, bone signals are also suppressed, which is useful for diagnosis tissue between fat and bone in knee images. However, since acquisition time of STIR MR is longer than other MR images, making STIR image is most effective to covert contrast MRI.
Methods
We conducted experiments in two stages. First, we confirmed whether deep learning is learning the bloch equation by phantom experiment and experimented whether it can be applied with in-vivo MRI images. PD, T1, and T2, which are components of the Bloch equation, are unique values of each tissue and can not be quantified for all tissues. Therefore, in order to consider all tissues in the phantom experiment, a total of 15,000,000 tissues were created using 100 PD from 0.01 to 1(0.01 interval), 500 T1 from 10ms to 5000ms(10ms interval), and 300 T2 from 10ms to 3000ms(10ms interval).
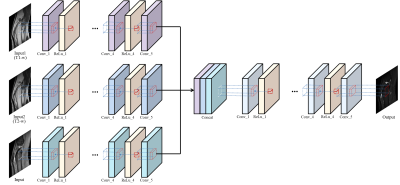
And we applied same method with in-vivo knee MRI. However, the in-vivo MRI does not follow the bloch equation exactly because there are many variables such as signal noise, hardware imperfection, B0, B1 inhomogeneity, and chemical shift and so on. Therefore, since information is insufficient only with T1 - and T2 - weighted images, we added a GRE image to give new information. That is, T1-, T2-weighted, GRE images were used as input images and STIR images were used as label images for learning. As shown in Figure 2, 5 convolution layers are used to extract features of each contrast image. Then, concatenated each features passes another 5 convolution layers to learn combination of features4. The total number of subjects was 12 and 8 slide images were used for each subject. The size of each image was 384×384 and 41×41 patch was used for learning.
Result and Discussion
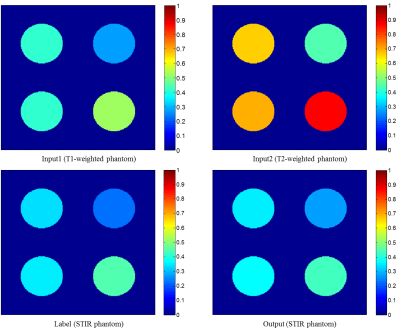
As shown in Figure 1, the phantom image shows that the STIR phantom image is perfectly created with T1- and T2-weighted phantom images. Through this phantom imaging experiment, we confirmed that deep learning completely learns the bloch equation and creates new contrast image.
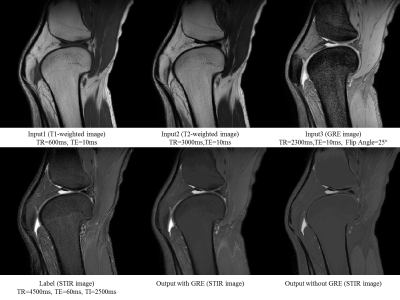
In Figure 3, bone and fat are all bright in T1-weighted and T2-weighted images, and it is not easy to observe the surrounding area. On the other hand, STIR images with suppressed fat components show that the signals of bones and fats are suppressed and darkened. Comparing the case of making an STIR image with only T1 - and T2 – weighted images and the case of adding a GRE image, it can be confirmed better when adding a GRE image on the contrast side or the detail side. This means that the GRE image is filling the missing information. Our output image suppressed fat and bone signals like STIR image, but does not make perfect to detail. Therefore, we need to find a way to learn without losing such detail information.
Conclusion
This study showed that the convolution neural network performs learning to make a new contrast STIR image with multi-contrast MRI. In addition, the simple bloch equation does not produce the variables that appear in the in-vivo image, whereas the deep learning can produce such a case well. It is confirmed that not only STIR images but also other contrast images can be generated.Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government(MSIP) (2016R1A2R4015016).References
[1]Bahrami, K., Shi, F., Rekik, I. & Shen, D. Convolutional neural network for reconstruction of 7T-like images from 3T MRI using appearance and anatomical features. In Proceedings of the 1st International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis (LABELS’16), 2016;39-47.
[2] Xiang, Lei, et al. "Deep Embedding Convolutional Neural Network for Synthesizing CT Image from T1-Weighted MR Image." arXiv preprint arXiv:1709.02073 (2017).
[3] Shanshan Wang. "Feasibility of Multi-contrast MR imaging via deep learning.", 25th ISMRM (2017)
[4] Wang, Limin, et al. "Places205-vggnet models for scene recognition." arXiv preprint arXiv:1508.01667 (2015).
Figures