2804
Reconstruction in deep learning of highly under-sampled T2-weighted image with T1- weighted image1School of Biomedical Engineering, Shanghai Jiao Tong University, shanghai, China, 2Department of Radiology and BRIC, University of North Carolina at Chapel Hill, chapel hill, NC, United States
Synopsis
T1-weighted image (T1WI) and T2-weighted image (T2WI) are routinely acquired in MRI protocols, which can provide complementary information to each other. However, the acquisition time for each sequence is non-trivial, making clinical MRI a slow and expensive procedure. With the purpose to shorten MRI acquisition time, we present a deep learning approach to reconstruct T2WI from T1WI and highly under-sampled T2WI. Our results demonstrate that the proposed method could achieve 8 or higher acceleration rate while keeping high image quality of the reconstructed T2WI.
Introduction
T1WI and T2WI are clinical routines for assessing structure and pathology, respectively. Typical scan time for both images is ~10 minutes, which also subjects to motion artifact from the patient. More patient groups will be able to be imaged if the scan time can be reduced further. Parallel imaging is commonly used to increase the acquisition speed by factors of 1.5 to 3 in most commercially available applications. In theory, much higher acceleration is possible but currently limited by artifact and signal-to-noise ratio (SNR) considerations. In this study, we propose a deep learning approach to achieve high acceleration rate of T2WI by incorporating T1WI into the reconstruction of the highly under-sampled T2WI. We adopt the deep fully convolutional neural network that consists of a contracting path and a symmetric expanding path that can leverage the context information from multi-scale feature maps. Our results suggest that the acceleration rate can be as high as 8 or above with negligible penalty of aliasing artifact and SNR.Methods
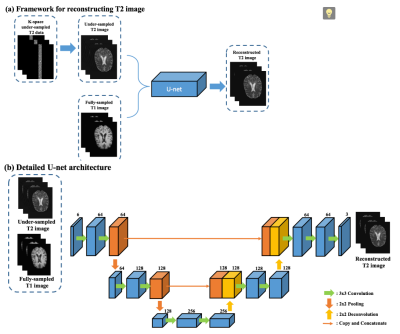
The framework for accelerating the T2WI reconstruction with T1WI and under-sampled T2WI is shown in Fig. 1(a). First, the original T2WI is retrospectively under-sampled and only the center k-space data with different ratios is kept to generate under-sampled T2WI. The under-sampled T2WI are concatenated with the fully-sampled T1WI, and then fed into the U-net neural network [1] for reconstructing the fully-sampled T2WI. To be specific, we implement the input as 6 consecutive axial slices (3 from fully-sampled T1WI and 3 from under-sampled T2WI). Thsee 6 slices are jointly considered as part of the feature maps since the first convolutional layer in the network. The output consists of 3 axial slices that correspond to the same position of the input. We synthesize every 3 consecutive axial slices for the reconstructed T2WI, and combine the outputs into the final 3D result by simple averaging. We regard this joint synthesis of the 3 consecutive slices as quasi-3D mapping. Detailed U-net architecture [1] has selected and been shown in Fig. 1 (b), as it has quite promising result in medical imaging field [2-4]. It consists of two paths: contracting path (on the left) and expansive path (on the right). We use convolution with zero-padding, so the final reconstructed T2WI has the same size as input images.Results
We utilized the dataset from the MICCAI Multiple Sclerosis (MS) segmentation challenge 2016 [5]. We selected 5 subjects with paired T1WI and T2WI. Peak signal-noise ratio (PSNR) and normalized mean squared error (NMSE) were used to quantitatively evaluate the reconstruction performance. The leave-one-out cross-validation strategy is employed for evaluation.
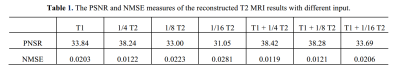
Average PSNR and NMSE have been summarized in Table 1. We explore performance of fully-sampled T1WI with different under-sampled T2WI ratios of 1/4, 1/8, 1/16. To illustrate the complementary effect between different contrasts, we also show results of the reconstructed T2WI with only T1WI. We can see that the image quality of ‘1/8 T2’ (PSNR: 33.00 dB) drops dramatically compared to ‘1/4 T2’ (PSNR: 38.24 dB), which implies that over under-sampled T2WI is difficult to be reconstructed. However, with the T1WI added, the reconstructed results of ‘T1 + 1/4 T2’ (PSNR: 38.42 dB) is even better than only using 1/4 T2WI. This verifies the idea that different contrasts have complementary information for each other. One interesting observation is that the ‘T1 + 1/8 T2’ (PSNR: 38.28 dB) is almost the same as ‘T1 + 1/4 T2’ (PSNR: 38.42 dB), which suggests that a high acceleration factor can be achieved with the proposed method.
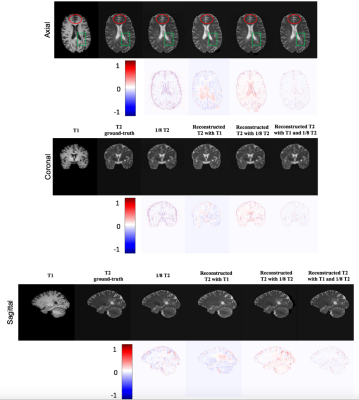
Visual observations for all comparisons are shown in Fig. 2. We find that the reconstructed T2WI using ‘1/8 under-sampled T2’ image has fuzzy boundary (red circle), but all the characteristics of T2 information in lesion is preserved (green box). On the other hand, the result of ‘predicted T2 with T1’ has clear tissue boundary but missing lesion information. So our proposed method combines T1 MRI and under-sampled T2 MRI as the input of U-net, which significantly improves the quality of reconstructed T2WI (last column).
Discuss and Conclusion
In this work, we propose a U-net architecture for reconstruction of high-quality T2WI with prior information of T1WI. The reconstruction is attained by a fast 3D image mapping with deep learning. Experimental results suggest superior performance of our method, including the perceptive quality of the outcomes and the high acceleration rate that could be as high as 8 or above. Although 2D acceleration was used in this work, this approach can be easily generalized to 3D case.Acknowledgements
No acknowledgement found.References
[1] O. Ronneberger, P. Fischer, and T. Brox, "U-net: Convolutional networks for biomedical image segmentation." pp. 234-241.
[2] C. M. Hyun, H. P. Kim, S. M. Lee et al., “Deep learning for undersampled MRI reconstruction,” arXiv preprint arXiv:1709.02576, 2017.
[3] X. Han, “MR‐based Synthetic CT Generation using a Deep Convolutional Neural Network Method,” Medical Physics, 2017.
[4] D. Lee, J. Yoo, and J. C. Ye, "Deep residual learning for compressed sensing MRI." pp. 15-18.
[5] O. Commowick, F. Cervenansky, and R. Ameli, "MSSEG Challenge Proceedings: Multiple Sclerosis Lesions Segmentation Challenge Using a Data Management and Processing Infrastructure."
Figures