2798
Iterative Cross-Domain Deep-Learning Approach for Reconstructing Undersampled Radial MRI1Yonsei University, Seoul, Republic of Korea
Synopsis
The purpose of this study is to eliminate the aliasing artifacts in accerelated radial MRI. We designed a Cross-Domain deep-learning network, called SISI-Net(Sinogram-Image-Sinogram-Image Network). This is an architecture to gradually solves data sparsity problems by iteratively learning the radial sampling data in the sinogram domain and the reconstructed data in the image domain. As a result, proposed network could remove aliasing artifacts effectively while maintaining structural information.
Introduction
Shortening MR acquisition time is one of the important areas of MR researches. To achive higher speed, various reconstruction methods have been developed for undersampled data. In particular, deep-learning based methods have recently been spotlighted, especially, iterative deep-learning is also being researched.1 Also, deep-learning can be efficiently performed on the sinogram domain obtained by 1D inverse fourier transform of radial k-space as well as the image domain.2 In this regard, we propose an iterative cross-domain CNN method that learns sinogram and image iteratively.
Methods
Radial MR can be used as a very effective acceleration method when combined with deep learning method. It is because that undersampled radial MR data can be efficiently interpolated in the sinogram domain through radon and iradon transform and is well trained for artifact removal through a deep-learning structure. To make training data set, we need complex process as in fig 1. First, the 2D-Fourier transform of the image is performed and then the 512 spokes radial data is generated by inverse gridding. Here, because it is common to use oversampling factor 2, 512 spoke is applied.3 Second, MR sinogram data were generated by 1D inverse fourier transform according to each spokes. Third, 1/8 undersampled 64 spokes radial MR data is interpolated through radon and iradon transform operations. Fig. 2 shows the architecture we have developed. It consists of two steps: first step is sinogram domain CNN (SCNN), which learns on the sinogram. Although SCNN is insufficient for artifact removal, but it uses raw data, so it has a characteristic that information loss is minimized. Second step is image domain CNN (ICNN), which learns on the image. This is specialized for artifact removal, but there is a disadvantage that the output image is blurred. Each CNN output of the cross-domain structure is uesd by the input of next CNN. We maximized performance by iteratively repeating the fig. 2 structure twice. We used 457 T2 fluid attenuated inversion recovery brain images(size=256*256) from Alzheimer’s Disease Neuroimaging Initiative MRI data.4 450 images were used for the train data, and 7 images were used for the test data. In each CNN layer, the ReLU transfer function is used to transfer the weight even at low gradient values, so that the fine learning can be performed as the learning in the SISI-Net structure becomes saturated. Patch size was 32*32, convolution filter size was 3*3, and the number of filter was 64.Results
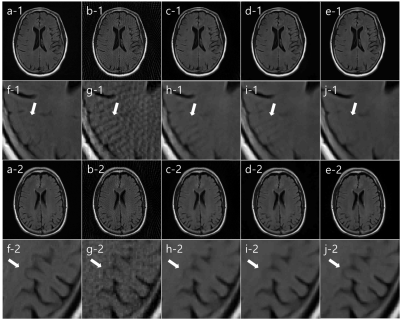
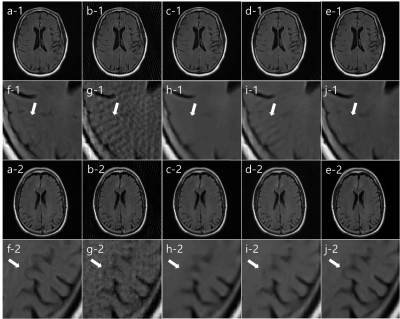
We compared our SISI-Net result with I-Net, KIKI-Net, R6_BM3D-mri, Wang’s methods.5, 6 In fig. 3, Reference image (a), undersampled radial MR image (b), I-Net (c), KIKI-Net (d), SISI-Net, our result (e), each magnified images (f-j) are shown. In both first and third row images, I-Net result has artifacts and cannot correct sufficiently. In image row 1, streak artifacts that did not exist in reference image (a), can be seen in KIKI-Net result (d) but not in SISI-Net result (e). This means that aliasing artifacts in the undersampled image (b) are misinterpreted as truly exists structures in KIKI-Net. In image row 3, we can see that the actual image structure is restored well in the SISI-Net result (e), but not in the KIKI-Net result (d). This can be interpreted that learning in the sinogram domain shares structural information with neighboring pixels in the convolution process, but k-space learning is not. In fig. 4, Reference image (a), undersampled radial MR image (b), R6-BM3D-mri method (c), Wang’s method (d), SISI-Net, our result (e), each magnified images (f-j) are shown. BM3D method shows removed streak artifact image but too much blurred. Wang’s method shows similar with I-Net result. Each average PSNR of 7 test images are (Underampled = 28.60, I-Net = 35.41, KIKI-Net = 36.58, R6_BM3D = 38.26, Wang’s = 32.50, SISI-Net = 39.40)Conclusion
We have developed a iterative deep learning architecture that improves image quality with only a few spokes radial MR data. In addition, it was confirmed that the result image is improved by repeatedly performing the sinogram domain and the image domain which is the reconstruction of the sinogram. The repetitive learning structure using sinogram can be used not only for radial MR but also for sparse-view undersampling of CT.Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No.2016R1A2B4015016).References
[1] Schlemper, Jo, et al. "A Deep Cascade of Convolutional Neural Networks for MR Image Reconstruction." International Conference on Information Processing in Medical Imaging. Springer, Cham, 2017. [2] Ye, Jong Chul, et al. "Projection reconstruction MR imaging using FOCUSS." Magnetic Resonance in Medicine 57.4 (2007): 764-775. [3] Beatty, Philip J., Dwight G. Nishimura, and John M. Pauly. "Rapid gridding reconstruction with a minimal oversampling ratio." IEEE transactions on medical imaging 24.6 (2005): 799-808. [4] ADNI data, ‘Alzheimer’s Disease Neuroimaging Initiative’ http://adni.loni.usc.edu/, accessed April, 2016 [5] Wang, Shanshan, et al. "Accelerating magnetic resonance imaging via deep learning." Biomedical Imaging (ISBI), 2016 IEEE 13th International Symposium on. IEEE, 2016. [6] Eksioglu, Ender M. "Decoupled algorithm for mri reconstruction using nonlocal block matching model: Bm3d-mri." Journal of Mathematical Imaging and Vision 56.3 (2016): 430-440.Figures