2786
Investigation of convolutional neural network based deep learning for cardiac imaging1Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, China, 2Department of Biomedical Engineering and Department of Electrical Engineering, The State University of New York, Buffalo, New York, Armenia
Synopsis
Deep learning based fast MR imaging has been very popular lately. Nevertheless, the empirical nature of existing approaches still leave quite a few questions open. To address this, this paper designs different convolutional neural networks to investigate various factors, such as direct CNN mapping, noise stimulation, data consistency and data sharing, for deep learning based cardiac imaging. We find out that if K-space manipulation strategy is not adopted, CNN still needs dedicated sampling patterns or more complicated structures to remove global corruptions. Furthermore, K-space updating strategy are encouraged to be incorporated with deep learning for better final performances.
Introduction
Applying deep learning to cardiac imaging is a very appealing direction and there have been preliminary results showing that convolutional neural networks (CNN) can empirically reconstruct MR images with improved quality compared to the classical approaches [1,2,3,4,5,6]. Nevertheless, the empirical nature of deep learning based cardiac imaging approaches still leave quite a few questions open, for example, whether CNN can really remove the global aliasing artifacts or not, how K-space update affects the final performances. Specifically, different from our own experience that CNN (especially shallow ones) is good at exploring local correlations rather than global correlations and may only perform well in local-corrupted sampling patterns such as 2D Poisson and 1D low frequency sampling masks [7,8], it was presented that the method developed in [6] has achieved exciting reconstruction results with 1D random Cartesian undersampling masks. To answer these questions, this paper designed five different models to study how different factors such as direct CNN mapping, noise stimulation, data consistency and data sharing affect the final performance of deep learning based cardiac imaging. The models have been tested on in vivo dataset and compared to the classical k-t FOCUSS method [9].Theory and method
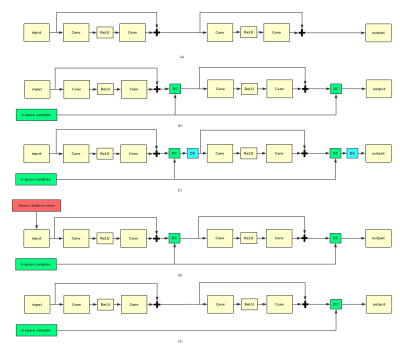
Five different CNNs have been designed to study different factors, where data consistency (DC) and data sharing (DS) are the K-space manipulation strategies, while direct CNN mapping and noise stimulation operate directly in the image domain. Specifically, DC projects back the original sampled k-space to enhance data fidelity. DS fills the entries using the samples from the adjacent frames to approximate missing k-space samples for cardiac imaging. Noise stimulation means we inject noise in the input to CNN with the purpose of augmenting the datasets and improving the robustness of neural networks. The basic CNN model adopts CNN to directly learn the nonlinear relationship between the undersampled and fully sampled cardiac MR images without K-space update. It has 3D convolution, ReLU activation and residual connection [10]. Model 1 is to test the performance of the direct CNN mapping, looking for an answer to whether CNN can remove the global aliasing artifacts or not. Model 2 added DC layers on top of model 1 [6] to check how K-space projection back for data-fidelity works for the final reconstructions. Model 3 add DS layer after each DC layer which explores the correlations between different frames of cardiac sequences and to check how this K-space correlation component contributes to the final results. Model 4 injects noise in the input to the network during training phase, trying to check whether noise disturbance can trigger CNN to remove the local correlation and therefore improve its final reconstruction performance. Model 5 adds a single DC layer to model 1 to check if only one time K-space update is enough for improving the final performances.Experiment
We collected 102 fully sampled cardiac MR data using 3T scanner (SIEMENS MAGNETOM Trio) with T1-weighted cine flash sequence. Multi-coil data were combined and then retrospectively undersampled using 1D random Cartesian masks [6]. After normalization and extraction, we got 17502 cardiac data, where 15000, 2000 and 502 were respectively used for training, validating and testing. Offline training of each model took almost 15 hours on a workstation (Intel Xeon (R) CPU E5-2680 V3 @2.5GHz *16, 128G).Results and discussion
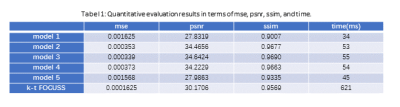
Fig. 2 presents the test results of different models and Table1 summarizes MSE, PSNR, SSIM values. Both visual and quantitative results show that CNN indeed can explore the temporal and spatial correlation of MR images and the reconstruction is very fast compared with k-t FOCUSS. However, from (d) and (e), we could observe shallow CNN settings can only remove limited global aliasing artifacts. Model 3 performs best and model 1 performs worst, which demonstrates that DC plays a key role in the encouraging performance of deep learning based MR imaging reconstruction methods and data sharing among different frames of cardiac imaging can also be helpful. In Table 1, model 4 is a little bit worse than model 2 and model 3, which means that noise stimulation could be disturbing for the final results. Model 5 doesn’t perform well either which again proves that K-space updating is important for the final reconstruction result.Conclusion
We have investigated how different neural network settings affect the final performances of deep learning based cardiac imaging with the finding that if K-space manipulation strategy is not adopted, CNN still needs dedicated sampling patterns or more complicated structures to remove global corruptions. Furthermore, K-space updating strategy are encouraged to be incorporated with deep learning neural networks for better final performances.Acknowledgements
Grant support: China NSFC 61471350, 61601450, the Natural Science Foundation of Guangdong 2015A020214019, 2015A030310314, the Basic Research Program of Shenzhen JCYJ20140610152828678, JCYJ20160531183834938, JCYJ20140610151856736 and the youth innovation project of SIAT under 201403 and US NIH R21EB020861 for Ying.References
[1] K. Kwon, D. Kim, H. Seo, J. Cho, B. Kim, H.W. Park, “Learning-based Reconstruction using Artificial Neural Network for Higher Acceleration”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2016, no. 24, p. 1801
[2] Y.S. Han, D. Lee, J. Yoo, Jong. Y, “Accelerated Projection Reconstruction MR Imaging using Deep Residual Learning”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2017, p. 0690
[3] S. Wang, Z. Su, L. Ying, X. Peng, S. Zhu, F. Liang, D. Feng, D. Liang, “Accelerating magnetic resonance imaging via deep learning”, ISBI 514-517 (2016)
[4] F. Knoll, “Leveraging the Potential of Neural Networks for Image Reconstruction”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2017
[5] B. Zhu, J. Liu, B. Rosen, M. Rosen, “Neural Network MR Image Reconstruction with AUTOMAP: Automated Transform by Manifold Approximation”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2017, p. 0640
[6] J. Schlemper, J. Caballero, J.V. Hajnal, A. Price, D. Rueckert, “A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction”, IEEE TMI, DOI: 10. 1109/TMI.2017.2760978 (2017)
[7] S. Wang, T. Xiao, S. Tan, Y. Liu, L. Ying, D. Liang, “Undersampling Trajectory Design for Fast MRI with Super-Resolution Convolutional Neural Network”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2017, p. 3984
[8] S. Wang, N. Huang, T. Zhao, Y. Yang, L. Ying, L. Dong, “1D Partial Fourier Parallel MR Imaging with Deep Convolutional Neural Network”, in Proceedings of the International Society of magnetic Resonance in Medicine (ISMRM), 2017, p. 0642
[9] H. Jung, J. C. Ye, and E. Y. Kim, “Improved k t BLAST and k t SENSE using FOCUSS,” Magnetic Resonance in Medicine, vol. 52, pp. 3201–3226, 2007
[10] He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
Figures