2782
A Neural Network for Referenceless Reconstruction in Simultaneous Multi-Slice Imaging1Fraunhofer MEVIS, Bremen, Germany, 2mediri GmbH, Heidelberg, Germany, 3University Bremen, Bremen, Germany
Synopsis
The unwrapping of simultaneous multi-slice images without extra reference data is presented. A trained deep neural network disentangles overlapping image content and creates the final magnitude images. The results are compared to established techniques (split slice-GRAPPA), especially where correct reference data are missing.
Introduction
Simultaneous multi-slice (SMS) imaging 1–3
has emerged as a promising acceleration technique for magnetic resonance
imaging (MRI) since multi-coil systems 4 and reconstruction methods like (split) slice-GRAPPA 5,6 paved the way for a variety of
applications. Established reconstruction strategies for SMS utilize spatial
encoding information inherent in multi-coil receiver arrays and require
additional reference data, i.e. auto-calibration signal (ACS), to disentangle
overlapping image content 7.
In general, ACS-acquisition can be time-consuming and a source of
reconstruction-errors.
Here, a deep neural network (DNN) was designed to unfold SMS images without the
need of any reference-scans. First, the DNN architecture is introduced. Thereafter,
the predicted images are evaluated and deep learning (DL) reconstructions (DLR)
are compared to split slice-GRAPPA (SSG) 6
where correct ACS is missing.
Methods
Training a DNN requires suitable
data. 37 datasets of phantom objects (N=30, fruits and geometric phantoms) and
heads (N=7) were used for training (Ntrain=29) and validation (Nval=8),
while evaluation was done on separate datasets of volunteers’ heads (Ntest=4).
All data were acquired with identical sequence parameters in 3 contrasts (TE=4.9/9.7/14.5ms,
TR=126ms, α=70°, matrix: 128x128x6). Rawdata of 20 receiver-coils were
preprocessed offline, i.e. simulated SMS acquisition and CAIPIRINHA-shifts 8,
and augmented by undersampling (90%,
80%, 75%, 70%, 60%, 50%, 40%, 30%) yielding to Ntrain=2349, Nval=648
datasets for a multiband factor (MB) of MB=2 and Ntrain=870, Nval=240
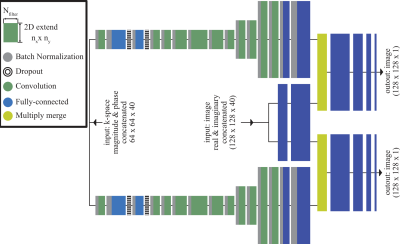
for MB=3, respectively. Figure 1 shows the DNN’s architecture where folded
(MB=2 or MB=3), uncombined, low-resolution k-space (64x64) and image data
(128x128) were fed into the network while sum-of-square combined single-band (SB)
images serve as target. SB-images were shifted to the according CAIPIRINHA-pattern.
Complex-valued input (Re, Imag) were concatenated along channel
dimension (Nch=40). A user defined loss-function: E = MSE x TV was applied to drive the optimizer.
Mean-squared error (MSE) and total-variation
error (TV) were combined to account
for global errors as well as mismatches on object borders. The DNN was set up
in the Keras library 9
and training (280 epochs in 28 hours) was performed on a Nvidia
GTX1080 graphics card.
All
images I of one dataset were normalized
to
$$I_n = \frac{I-I_{min}}{I_{max}- I_{min} } \text{ .}$$
Two metrics were introduced for quantification of the differences between reconstructions. The commonly used normalized root mean-squared error (NRMSE) together with subtraction-maps of SB and recovered SMS-images and the structural similarity index (SSIM) 10. SSIM assesses perceptual image quality by taking advantage of characteristics of the human visual system 10. It is defined as
$$SSIM(x,y) = L(x,y) \cdot C(x,y) \cdot S(x,y) \text{ ,}$$Abstract
where L , C , S are luminance, contrast and structure for each pixel (x,y) 10.
For NRMSE and mean SSIM (MSSIM) a threshold-mask of 1% was applied to
remove noise outside the object.
Results
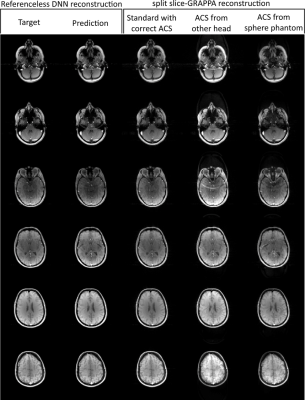
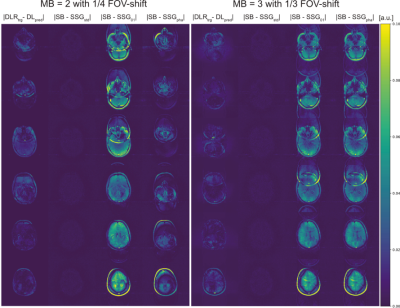
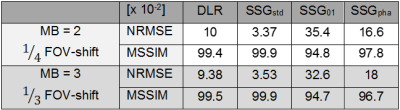
The different reconstruction results are shown in Figure 2. Unseen test data were fed into the trained DNN and its predictions were evaluated and compared to SSG reconstructions with suitable ACS (SSGstd), ACS taken from another object with similar anatomy, i.e. another head, (SSG01) and ACS of a spherical phantom (SSGpha). All measurements had an identical field-of-view (FOV).Normalized subtraction-maps (Fig.3) compare DNN's prediction to standard SSG and SSG where correct ACS are missing and replaced by other ACS (SSG01, SSGpha). SMS-reconstructions for MB=2 with 1/4 FOV-shift (left) and MB=3 (1/3 FOV-shift) (right) are illustrated. Both settings are quantified by NRMSE as tabulated in Table 1.
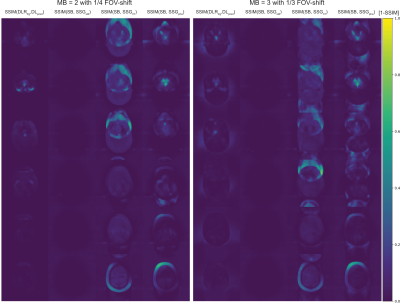
Figure 4 depicts the inverse SSIM-maps (1-SSIM) for the above cases. The corresponding MSSIM are given in Table 1.
Discussion
Visual inspection generally approves the unwrapping of SMS data by a DNN (Fig.2). Although more inter-slice signal leaks into the reconstructed slices, DLR outperforms SSG where ACS data are missing or corrupted. SSIM allows more perceptual than purely pixel-wise evaluation of the different approaches and emphasizes the above impression. Nevertheless, reconstruction errors especially inside the region of interest cannot clearly be distinguished from pathological findings at this stage. The limited amount of trainings data and the broad spectrum of these (fruits, phantoms, heads) may have prevented more efficient training. As in other DL scenarios, it is likely that the performance can be significantly increase when additional training data are included, ideally these would be provided by a large database with in-vivo raw-data.Conclusion
A novel approach to reconstruct SMS images without additional ACS is presented. Although, clearly more reconstruction artifacts as for a correct SSG reconstruction occur, DLR is beneficial to SSG with incorrect reference data. This might be especially useful for dynamic imaging in presents of motion or for emergency-protocols as no ACS is needed and demanded computational load is low.Acknowledgements
The authors gratefully thank Markus Wenzel and Hans Meine for valuable, interdisciplinary discussions on deep learning in image-processing.References
1. Weaver
JB. Simultaneous multislice acquisition of MR images. Magn Reson Med 1988;8:275–284.
2. Müller S.
Multifrequency selective rf pulses for multislice MR imaging. Magn Reson Med
1988;6:364–371.
3. Souza SP.
Simultaneous Multislice Acquisition of MR Images by Hadamard-Encodes Excitation
SOUZA_1988.pdf.
4. Larkman
DJ, Hajnal J V., Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil
arrays for separation of signal from multiple slices simultaneously excited. J
Magn Reson Imaging 2001;13:313–317.
5. Setsompop
K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled
aliasing in parallel imaging for simultaneous multislice echo planar imaging
with reduced g-factor penalty. Magn Reson Med 2012;67:1210–1224.
6. Cauley
SF, Polimeni JR, Bhat H, Wald LL, Setsompop K. Interslice leakage artifact
reduction technique for simultaneous multislice acquisitions. Magn Reson Med
2014;72:93–102.
7. Barth M,
Breuer F, Koopmans PJ, Norris DG, Poser BA. Simultaneous multislice (SMS)
imaging techniques. Magn Reson Med 2016;75:63–81.
8. Breuer
FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM. Controlled
aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for
multi-slice imaging. Magn Reson Med 2005;53:684–691.
9. Chollet F
et al. Keras. 2015.
10. Wang Z,
Bovik AC, Sheikh HR, Member S, Simoncelli EP, Member S. Image Quality
Assessment: From Error Visibility to Structural Similarity. 2004;13:600–612.
10. Wang Z, Bovik AC, Sheikh HR, Member S, Simoncelli EP,
Member S. Image Quality Assessment: From Error Visibility to Structural
Similarity. 2004;13:600–612.
Figures