2773
Fat Content and Fatty Acid Composition Quantification Using a 3D Stack-of-Radial Trajectory With Adaptive Gradient Calibration1Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 3MR R&D Collaborations, Siemens Healthineers, Cary, NC, United States, 4Radiology, Duke University Medical Center, Durham, NC, United States, 5Center for Advanced Magnetic Resonance Development, Duke University Medical Center, Durham, NC, United States
Synopsis
The purpose of this study was to evaluate the effect of an adaptive gradient calibration technique for a 3D stack-of-radial sequence with regard to magnitude- and complex-based fat content quantification and triglyceride saturation estimation. In-vivo measurements in two healthy volunteers showed that gradient calibration improved the accuracy of complex fitted fat fraction and fatty acid maps. Gradient calibration only had a minor impact on magnitude-based fat fraction results.
Introduction
Several Cartesian multi-echo MRI methods for the quantification of liver fat content1,2 and fatty acid composition (FAC)3-5 are available. Common for all Cartesian trajectories is that the same readout direction is applied for all acquisitions. Thus, gradient imperfections and delays as well as eddy current errors can either be neglected (for unipolar readout gradients)1,3, or modeled as a phase discrepancy between even and odd echoes (for bipolar readout gradients)2,4,5. Compensating for gradient delays in radial trajectories can be done adaptively6, but when jointly performed for all receive channels this does not address higher-order phase variations as addressed in Cartesian approaches. For multi-echo acquisitions, an adaptive gradient calibration (GC) procedure was recently proposed that compensates for the effective gradient delay for individual receive channels and echoes7. However, the impact of such a technique on magnitude-based fat estimation and on FAC quantification remains unexplored. The purpose of this study was to evaluate the effect of an adaptive GC method on complex- and magnitude-based proton density fat fraction (PDFF) and FAC quantification.Methods
Gradient calibration: An adaptive gradient calibration procedure6,7 that compensates for eddy-current-induced gradient delays and deficiencies was developed for a bipolar, 3D stack-of-radial sequence. The procedure acquired calibration spokes prior to the actual image acquisition, averaged them across all partitions, and then calculated shifts in k-space for every channel and echo. The estimated shifts were used to compensate gradient errors in subsequently acquired k-space lines by shifting them along the readout direction. The gradient calibration was implemented in C++ and integrated into the scanner reconstruction pipeline.
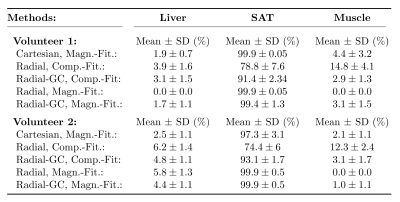
PDFF estimation: Bipolar, multi-echo data from two healthy volunteers (1 male, 1 female) was acquired with a prototypical 3D stack-of-radial trajectory (12 echoes, TE1 = 1.27ms, ΔTE = 1.21ms, TR = 17ms, voxel size = 1.98x1.98x5mm3, bandwidth = 1185Hz/px, FA = 4°) using an 18-channel body and a spine array coil on a 3T MR system (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) during free-breathing. Every experiment was reconstructed twice: once without calibration (termed "Radial"), and once including the described gradient calibration procedure (termed "Radial-GC"). PDFF map estimation for the Radial and Radial-GC scans was performed using both complex-8 and magnitude-based9 parameter fitting of the multi-echo image series, respectively (termed "Complex-Fitting" and "Magnitude-Fitting"). A time-domain calibration of the fat signal dephasing optimized for liver applications was used10. The methods were compared with PDFF values from a prototypical Cartesian 3D VIBE (6 echoes, TE1 = 0.94ms, ΔTE = 0.88ms, TR = 7ms, voxel size = 2.5x2.5x3mm3, bandwidth = 1565Hz/px, FA = 4°) acquisition. Quantitative evaluation was based on mean and standard deviation of PDFF values within regions of interests (ROIs) in the liver, muscle and subcutaneous adipose tissue (SAT).
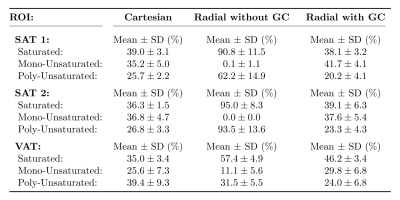
Fatty acid composition: In one of the volunteers, an additional scan in the lower abdomen was acquired using the described Radial and Radial-GC scans. A 9-peak fat model11 with variable peak amplitudes was applied and described using the number of double bonds (ndb) and number of methylene-interrupted double bonds (nmidb)11,12. Other than that, the same signal model and protocol settings as for PDFF calculation were used. The parameters ndb and nmidb were calculated by fitting the denoised13,5, complex images to the non-linear model, and then maps of the saturated, mono-unsaturated and poly-unsaturated fat were determined3. Additionally, bipolar, 12-point multi-echo data using a prototypical GRE sequence (2D multi-slice, 12 slices, TE1 = 1.27ms, ΔTE = 1.23ms, TR = 193ms, FA = 10°, bandwidth = 1955Hz/px, voxel size = 3.6x3.6x10.0mm3) was acquired and processed by means of Cartesian FAC quantification5. Fatty acid values within ROIs in the SAT and visceral adipose tissue (VAT) were compared for all reconstructions.
Results and Discussion
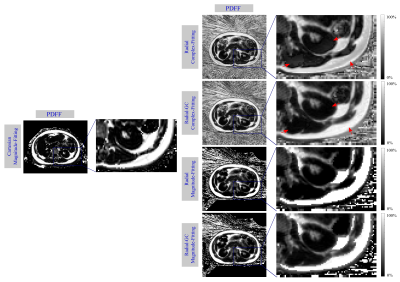
Fig. 1 depicts exemplary PDFF maps, and Table 1 illustrates the quantitative PDFF evaluation. For the complex-fitting technique, GC improved the visual impression of the PDFF map and the accuracy in the liver, muscle and SAT, making it more consistent with the PDFF results of the Cartesian reference. For the magnitude-fitting approach, GC had only small effects on the estimated PDFF values.
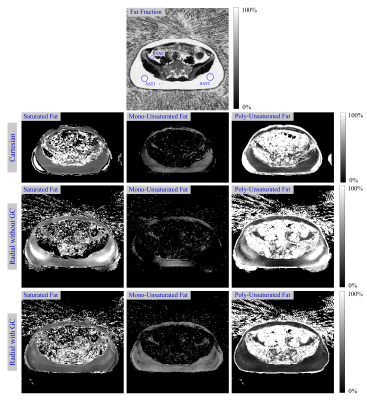
Fig. 2 shows the calculated in-vivo FAC maps, and Table 2 depicts respective ROI measurements. Radial MRI without GC failed to reliably estimate in-vivo fatty acid maps, especially for the mono-unsaturated component. The adaptive GC technique improved the accuracy of the radial method with regard to the Cartesian reference method, in particular in the SAT.
Conclusion
The adaptive GC method improved the accuracy of complex-based PDFF estimation and FAC quantification techniques with regard to Cartesian reference methods. GC had only small effects on magnitude-based PDFF maps.Acknowledgements
No acknowledgement found.References
1. Yu H, McKenzie CA, Shimakawa A, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26(4):1153-1161.
2. Peterson P, Månsson S. Fat quantification using multiecho sequences with bipolar gradients: investigation of accuracy and noise performance. Magn Reson Med. 2014;71(1):219-229.
3. Peterson P, Månsson S. Simultaneous quantification of fat content and fatty acid composition using MR imaging. Magn Reson Med. 2013;69(3):688-697
4. Leporq B, Lambert SA, Ronot M, et al. Hepatic fat fraction and visceral adipose tissue fatty acid composition in mice: Quantification with 7.0 T MRI. Magn Reson Med. 2016;76(2):510-518.
5. Schneider M, Lugauer F, Nickel D, et al. Improving the Noise Propagation Behavior of Different Fatty Acid Quantification Techniques using Spectral Denoising. In: Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, HI, USA; 2017. p. 5195.
6. Block K, Uecker M. Simple method for adaptive gradient-delay compensation in radial MRI. In: Proceedings of the 19th Annual Meeting of ISMRM, Montreal, Canada; 2011. p. 2816.
7. Armstrong T, Dregely I, Stemmer A, et al. Free-breathing liver fat quantification using a multiecho 3D stack-of-radial technique. Magn Reson Med. 2017. http://dx.doi.org/10.1002/mrm.26693.
8. Nickel MD, Zhong X, Kannengiesser S. Water-fat separation using low-resolution phase maps and eddy current compensation. In: Proceedings of the 30th Annual Scientific Meeting of ESMRMB, Toulouse, France; 2013. #607.
9. Zhong X, Nickel MD, Kannengiesser SA, et al. Liver fat quantification using a multi-step adaptive fitting approach with multi-echo GRE imaging. Magnetic resonance in medicine. 2014;72(5):1353-1365.
10. Nickel MD, Kannengiesser SAR, Kiefer B. Time-domain calibration of fat signal dephasing from multi-echo STEAM spectroscopy for multi-gradient-echo imaging based fat quantification. In: Proceedings of the 23rd Annual Meeting of ISMRM, Toronto; 2015. #3658.
11. Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat 1H MR spectrum. NMR Biomed. 2011;24(7):784-790.
12. Bydder M, Girard O, Hamilton G. Mapping the double bonds in triglycerides. Magn Reson Imaging. 2011;29(8):1041-1046.
13. Lugauer F, Nickel D, Wetzl J, et al. Robust Spectral Denoising for Water-Fat Separation in Magnetic Resonance Imaging. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. p. 667-674.
Figures