2758
A novel strategy for rapid multiparameter mapping based on SPGR with continuous steady state longitudinal magnetization1Department of Biomedical Sciences, Seoul National University, Seoul, Republic of Korea, 2Department of Radiology, Seoul National University, Seoul, Republic of Korea
Synopsis
A method is proposed for simultaneous T1, T2* and M0 mapping on a single scan by removal of inter-scan time delays based on the analytically found arrays of flip angles and TRs that maintain longitudinal magnetization in a steady state throughout the scan. Our preliminary results are in support of potential application of the proposed method in rapid multiparametric MRI in combination with a suitable undersamping strategy.
Introduction
Rapid multiparametric MRI has been an important issue, and several promising methods have been reported1,2,3. While a single scan method2,3 is preferred, multiple scans4,5,6 may be necessary for fully sampled data acquisition4,5,6 by repeating the same sequence (e.g., linear Cartesian4,5 and interleaved spiral6 encoding) or for additional image contrast1 by using different sequences (e.g., SPGR+bSSFP1). In either ways, there exist inter-scan time delays which may substantially increase the total scan time. In this report, we propose a data acquisition and post-processing strategy for simultaneous T1, T2*, and M0 mapping on a single-scan per slice using SPGR with Cartesian encoding. The proposed single-scan multiparameter mapping is achieved by removal of inter-scan time delays by maintaining longitudinal magnetization in a steady state throughout the scan based on analytically found arrays of TR and flip angle (FA).Methods
Theory
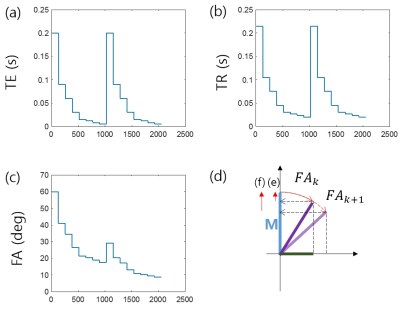
The arrays of FAs and TRs for maintaining the longitudinal magnetization throughout the scan group (Fig.1) can be found analytically by geometrical consideration shown in Fig.1(d). Based on the calculated arrays of FAs and TRs, a scheme for simultaneous T1, T2*, and M0 mapping on a single-scan can be designed as in Fig.1. The whole scan consists of two groups of scans with no time delay in-between (Fig.1). The same arrays of TEs and TRs are used for the scan groups 1 and 2 but with different arrays of FAs resulting from the different initial FAs of 60.0˚ and 29.1˚. The eight steps in each scan group result from the 8-point T2* mapping.
T2* and M are calculated by exponential fitting (Eq.1). T1 and M0 can be calculated from the two different steady state magnetizations M1 and M2 from the scan groups 1 and 2 as in the DESPOT1 method1 with an optional B1 mapping. Thus, two solutions for T2* are available from M1 and M2, and three solutions for M0 from M1, M2, and DESPOT1. They are averaged to yield a final set of parameter maps.
$$$S_{k}=M\sin{FA_{k}}e^{-\frac{TE_{k}}{T2^{*}}}$$$ [1]
Experiments
Experimental data were acquired at 9.4T (single-channel surface coil (Agilent)). Matlab (Mathworks Inc.) was used for data post-processing. The proposed method was tested on five phantoms (2 cc vials) with different MnCl2 concentrations4,7 and on a tomato.
The following sequence parameters were used for the evaluation of the proposed method: TE=200/90/60/30/15/12/8/5 ms, TR=TE+15 ms, FA=60.0/40.9/34.3/26.4/21.5/20.4/18.8/17.5 ˚ for the scan-group 1 and 29.1/20.2/17.1/13.2/10.8/10.2/9.4/8.8 ˚ for the scan-group 2. The gradient moment of the spoiler was 6600 mTms/m. The total scan time was 138 s.
As a reference for comparison, T1, T2*, and M0 of the phantoms were estimated using the following methods: T1 (spin-echo; 10 TIs=10/20/40/84/172/349/711/1450/2900/6000 ms; TR=10000 ms), M0 and T2* (multi-echo SPGR; ETL=8, minimum TEs=30/5/2.5/2.5/2.5 ms and echo-spacings =30/10/8/4/3 ms for phantoms #1-#5; TR=2000 ms). DESPOT1 (FA=13/64 ˚, TE=3 ms, TR=200 ms) and multi-echo SPGR (ETL=8, minimum TE=2.5 ms, echo-spacing=12 ms) were used for T1, T2*, and M0 mapping of the tomato. For the optional B1 mapping, SPGR was used (FA= 60/120 ˚, TR=2000 ms, TE=10 ms).
Results
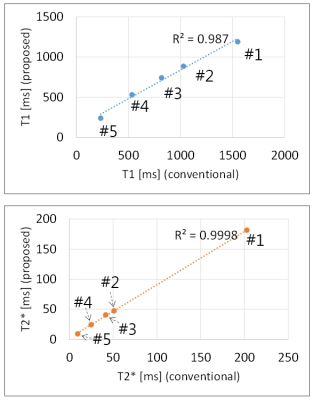
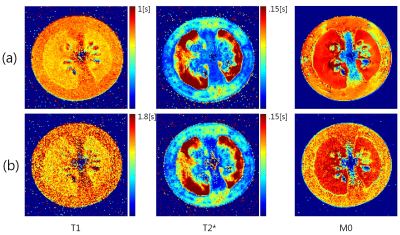
The T1 and T2* of the phantoms estimated by the conventional and proposed methods are compared in Fig.2. An R2 of 0.9870 was obtained for T1 with a trend toward underestimation in the long T1 range (phantoms #1-#2). An R2 of 0.9998 was obtained for T2* with a slight underestimation for the phantom #1 (longest T2*). The T1, T2*, and M0 maps of the tomato are in close agreement except for the higher noise level with the proposed method (Fig.3).Discussion
The proposed method allows for rapid multiparameter mapping on a single-scan per slice by removal of inter-scan time delays, which was achieved by maintaining the steady state longitudinal magnetization. The accuracy of the proposed method for long T2* may be improved by optimizing TEs and spoiling. Incomplete spoiling would be manifested as an artifact in the long T2* regime. The higher SNR of the reference parameter maps may be due mainly to the sequence parameters used (e.g., long TR, large FA). The trend toward underestimated T1 in the long T1 range requires further investigation particularly on the FA optimization as for DESPOT11.
Despite these several limitations, our preliminary results are in support of the potential application of the proposed method in rapid multiparametric MRI upon further reduction of scan time by referring to the existing undersampling methods such as key-hole and compressed sensing.
Conclusion
Upon further optimization of TEs, FAs, spoiling, and scan time in particular, the proposed method may be used for rapid multiparametric MRI.Acknowledgements
This work was supported by the National Research Foundation funded by the Korean government, MSIP (NRF-2014M3A9B6069340) and the Ministry of Education, Science and Technology (2016R1D1A1B03931233) of Korea.References
1. Deoni SCL, Rutt BK, and Peters TM. Rapid Combined T1 and T2 Mapping Using Gradient Recalled Acquisition in the Steady State, Magn Reson Med. 2003;49:515-526.
2. Warntes JBM, Dahlqvist O, and Lundberg P. Novel Method for Rapid Simultaneous T1, T2*, and Proton Density Quantification, Magn Reson Med. 2007;57:528-537.
3. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting, Nature 2013;495:187-193.
4. Gao Y, Chen Y, Ma D, et al. Preclinical MR fingerprinting (MRF) at 7 T: effective quantitative imaging for rodent disease models, NMR Biomed. 2015;28:384-394.
5. Anderson CE, Wang CY, Gu Y, et al. MR fingerprinting (RIPE-MRF) for enhanced motion artifact suppression in preclinical Cartesian MR fingerprinting, Magn Reson Med. 2017; DOI: 10.1002/mrm.26865
6. Jiang Y, Ma D, Seiberlich N, et al. MR Fingerprinting Using Fast Imaging with Steady State Precession (FISP) with Spiral Readout, Magn Reson Med. 2015;74:1621-31.
7. Ben-Eliezer N, Sodickson DK, Block KT. Rapid and Accurate T2 Mapping from Multi-Spin-Echo Data Using Bloch-Simulation-Based Reconstruction, Magn Reson Med. 2015;73:809-817.
Figures