2743
Optimization of Asymmetric Spin Echo MRI for Oxygen Extraction Fraction Mapping in the Brain and Initial Experience with Moya-Moya Patients1Radiology, Florida Hospital & University of Central Florida School of Medicine, Orlando, FL, United States, 2Mallinckrodt Institute of Radiology, Washington University School of Medicine, St. Louis, MO, United States, 3Washington University School of Medicine, St. Louis, MO, United States, 4University of Iowa School of Medicine, Iowa City, IA, United States
Synopsis
Asymmetric Spin Echo (ASE) MRI has been previously applied for quantitative cerebral oxygen extraction mapping. In this study we optimize this technique using O-15 PET as a gold standard for oxygen extraction fraction (OEF) quantitation and apply the optimized ASE approach for studying brain lesions in Moya-Moya patients. Results suggest that optimized OEF maps from ASE MRI have the potential to detect brain lesions unseen with conventional MRI sequences. These lesions detected by ASE appear to provide information that is statistically independent from the information provided by conventional MRI approaches.
Introduction
Non-invasive, rapid, and high-resolution means of mapping oxygen extraction and metabolism in the brain is key to early detection of lesions prior to their evolution to edema or gliosis which can eventually be detected with conventional MRI methods. Towards this means, Asymmetric Spin Echo (ASE) MRI has been previously applied for quantitative cerebral oxygen extraction mapping1 using the model developed by Yablonskiy and colleagues2. The objective of this study is to optimize this technique using O-15 PET as a gold standard for oxygen extraction fraction (OEF) quantitation and apply the optimized ASE approach for studying brain lesions in Moya-Moya patients.Methods
ASE Echo Planar Imaging and O-15 PET imaging of the brain were performed on six Moya-Moya patients in accordance with an IRB-approved protocol using the previously established MRI sequence1 (imaging parameters: TR = 5 sec, TE = 62 ms, FOV = 192 x 192 mm2, matrix size = 64 x 64, slice thickness = 3 mm, and EPI readout bandwidth = 160 kHz) and standard clinical O-15 PET. Conventional DWI, GRE, FLAIR, and T1 and T2 weighted MRI of the brain was also performed along with the ASE imaging. PET and MRI data were co-registered using the Montreal Neurological Institute Standard Atlas method. OEF maps were computed for all six patients from both their MRI and the PET data. An iterative neural network was implemented on the Center for High Performance Computing’s (CHPC) supercomputer at Washington University to learn the optimal ASE model parameters that allow the ASE sequence to yield OEF maps that best match the O-15 PET OEF maps. Using these optimized parameters, OEF maps were recomputed from the ASE data.
In an effort to understand whether or not the brain lesions detected in the Moya-Moya patients by the optimized ASE OEF maps provide information that is statistically independent from the information provided by conventional clinical MRI sequences, independent component analysis was performed on the ASE OEF and conventional MRI data using a previously established algorithm3 implemented on the CHPC supercomputer on Matlab.
Results & Discussion
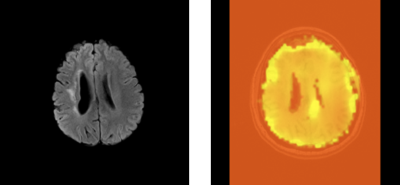
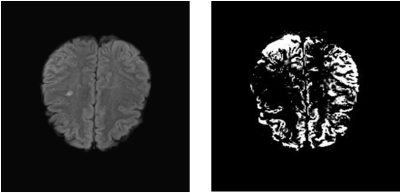
Prior to neural network based optimization, the EOF maps generated from ASE data did not reliably mimic the PET-OEF calculations. Upon iterative optimization, a close match between these two mapping paradigms was obtained for all six patients. Using the optimized modelling parameters for ASE, OEF maps were computed that clearly demonstrated brain lesions in Moya-Moya patients that could not be detected with conventional MRI sequences. Out of the conventional MRI sequences, FLAIR was best able to demonstrate lesions that could be detected without ASE. An example of such a lesion is shown in Figure 1 for one of the Moya-Moya patients, in whom FLAIR and the remaining convention sequences could not detect the left medial peritrigonal lesion. Similarly, the primary independent components of the MRI data demonstrated lesions seen with the optimized ASE technique that were not seen with FLAIR and other conventional sequences. An example of such a lesion is shown in Figure 2. This lesion is hence statistically independent, i.e. contains information that is distinct from what conventional MRI can provide.
Conclusion
In conclusion, optimized OEF maps from ASE MRI have the potential to detect brain lesions unseen by conventional MRI sequences. The additional lesions detected by ASE appear to provide information that is distinct from that provided by conventional MRI approaches. This distinction may be useful for early detection of brain lesions that arise due alteration in oxygen extraction/metabolism before edema or gliosis occurs.Acknowledgements
The authors acknowledge the staff at the Center for High Performance Computing at Washington University for their support in running complex batch processes.
Study funding: Supported by NINDS RO1 NS051631 (CPD). Support for the research imaging center where the MR studies were performed was provided by 1 UL1 RR024992-01, 1 TL1 RR024995-01 and 1 KL2 RR 024994-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), NIH Roadmap for Medical Research and P30NS048056.
References
1. An, H. et al. (2003) MRM 50: 708-716.
2. Yablonskiy DA and Haacke EM, MRM 32:749-763, 1994.
3. Tailor et al, Vision Research 40(19): 2671-6, 2000
Figures