2735
U-net: Convolutional Networks for Carotid Artery Wall Segmentation in Simultaneous Non-Contrast Angiography and intra-Plaque hemorrhage (SNAP) imaging1Department of Electronic Engineering, City University of Hong Kong, Hong Kong, Hong Kong, 2Center of Biomedical Imaging Research, Tsinghua University, Beijing, China, 3Beijing Jiaotong University, Beijing, China, 4Beijing Tian Tan Hospital, Beijing, China
Synopsis
The purpose of this study is to develop a U-net deep learning model to segment the carotid artery wall using a single 3D Simultaneous Non-Contrast Angiography and intra-Plaque hemorrhage (SNAP) acquisition. Using U-net convolutional Networks can achieve acceptable dice similarity coefficient. In addition, by adding more SNAP imaging such as phase-corrected images (CR), the magnitude of REF and the real part of IR as well as excluded the slice that cannot register and has low image quality may further improve the result.
Introduction
Stroke is one of the commonly leading causes of death and disability worldwide[1]. Carotid artery atherosclerosis is a main cause of stroke [2]. In order to evaluate the risk of carotid atherosclerotic plaque, black-blood vessel wall MRI techniques has been proposed to assess the plaque burden and plaque components[2]. However, multiple contrast are needed, which are challenge to analysis. Recently, a Simultaneous Non-Contrast Angiography and intra-Plaque hemorrhage (SNAP) technique has been proposed[3] and studies has demonstrate its ability to identify luminal stenosis[3] and plaque components[4, 5] with a single scan. However, the ability of SNAP to quantify the plaque burden has not been proven. Thus, the aim of this study is to develop an automatic technique to delineate the vessel wall on SNAP.Methods
In this study, the SNAP images with the lumen contours labeled in previous published population[5] were included. In this dataset, the lumen and outer wall contours were drawn on traditional multi-contrast images[6], such as TOF, T1W and T2W. Then the contours were mapped to the corresponding re-sliced SNAP images. In the multiple images SNAP can generate, we choose the magnitude of the inversion recovery acquisition (IR) to carry out carotid artery wall segmentation. As a pre-processing, the images were resampled to 0.3125*0.3125mm2 in-plane resolution and smoothed by Gaussian filter. In this study, a U-net convolutional networks[7] was used to train the labeled data, and classify pixels and get the wall region at last (the pixel belongs to wall region labeled as 1 and other labeled as 0). We keep the input and output shape of the convolutional layer consistent via padding zeros. Statistical analysis was performed: compare the wall thickness between the manual segmentation and our algorithm and their statistical significantly, calculated Dice Similarity Coefficient (DSC) for the proposed algorithm.Result
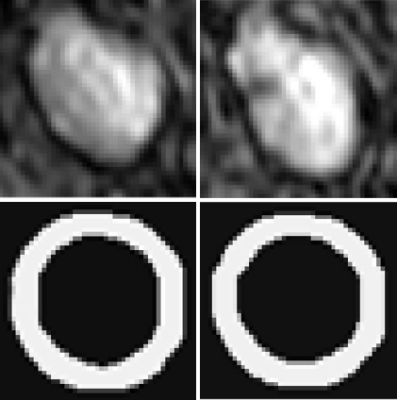
2059 slices were included in this study, 1837 slices were used for training, 222 slices for testing. Fig. 1 shows examples of the segmentation result of the proposed method. The proposed method can have a similar segmentation results comparing with the manual segmentation. Maximum mean carotid artery wall thickness (CAWT) measure by our proposed algorithm was 1.79$$$\pm$$$0.39mm, while the manual carotid artery wall thickness measure was 1.64$$$\pm$$$0.51 mm, the t-test showed there is no significant difference between the results of proposed algorithm and manual segmentation (P=0.0042) (Table 1). The pixel wise dice similarity coefficient reaches 73.72%, which means 73.72% of carotid wall region identified by proposed algorithm and manual segmentation was overlap (Table 2).Discussion and Conclusion
In this study, a U-net Convolutional Networks was used to segment the carotid wall in Simultaneous Non-Contrast Angiography and intra-Plaque hemorrhage (SNAP) images. Only magnitude of IR generated from SNAP imaging was used and the results were acceptable, which means the proposed algorithm has potential to segment the carotid wall. In the future, we should consider adding more SNAP images in such as phase-corrected images (CR), the REF, et al. Because these images were natural co-registered, add them should be straight-forward. Also, the final segmentation is determined by simply thresholding the output of CNN, which may generate rough segmentation. Utilizing the active contour or level set methods to determine the final segmentation contours should be tested in future studies.Acknowledgements
No acknowledgement found.References
[1] Mackay J, Mensah GA (2004) The atlas of heart disease and stroke. World Health Organization, Geneva
[2] A. Gupta, H. Baradaran, A. D. Schweitzer, H. Kamel, A. Pandya, D. Delgado, A. Dunning, A. I. Mushlin, and P. C. Sanelli, “Carotid plaque mri and stroke risk a systematic review and meta-analysis,” Stroke, vol. 44, no. 11, pp. 3071–3077, 2013.
[3] Wang, Jinnan, et al. "Simultaneous noncontrast angiography and intraplaque hemorrhage (SNAP) imaging for carotid atherosclerotic disease evaluation." Magnetic resonance in medicine 69.2 (2013): 337-345.
[4] Chen, S., et al., Carotid Atherosclerotic Plaque Surface Condition Evaluation Utilizing Simultaneous non-contrast Angiography and intra-Plaque hemorrhage (SNAP) Sequence, in International Society for Magnetic Resonance in Medicine. 2015.
[5] Qiang Zhang et al. Automatically Identify Plaque Components in Carotid Artery using Simultaneous Non-Contrast Angiography and intraPlaque hemorrhage (SNAP) imaging, ISMRM 25th Annual Meeting & Exhibition
[6] Saam, T., et al., Quantitative evaluation of carotid plaque composition by in vivo MRI. Arterioscler Thromb Vasc Biol, 2005. 25(1): p. 234-9.
[7] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2015.