2734
Automatic Myocardium Segmentation using Fully Conventional Network (FCN)1University of California, San Francisco, San Francisco, CA, United States
Synopsis
We introduce a new methodology that combines deep learning and level set for the automated segmentation of the myocardium from cardiac cine magnetic resonance (MR) data. The method employs deep learning algorithm to learn the segmentation task from the ground truth data. The inferred shape is incorporated into
Purpose
Automatic segmentation provides essential and efficient tools for accurate functional measurements, especially manual segmentation is impractical for analyzing massive and complex 3D/4D datasets. We here propose an automatic segmentation based on deep learning and level set methods to efficiently segment myocardium from 3D cardiac cine MRI data.Methods
Data acquisition: A highly accelerated free-breathing self-gated 3D cardiac cine MRI [1] was applied on 10 healthy volunteers (5 females, 5 males, age 32.4±14.6 years and heart rate = 64.8±8.8 bpm) on a 3.0T MR scanner (GE Medical Systems, Milwaukee, WI) with an 8-channel cardiac coil. Scan parameters: FOV=34.0×25.5cm2, TR/TE=4.1/1.7ms, FA=60°, BW=±125kHz, slice thickness of 4mm, image matrix=256×144, temporal resolution of 41 ms, and scan time of 2.5±0.3 minutes. Images were reconstructed using a combined compressed sensing and parallel imaging method, k-t SPARSE-SENSE [2].
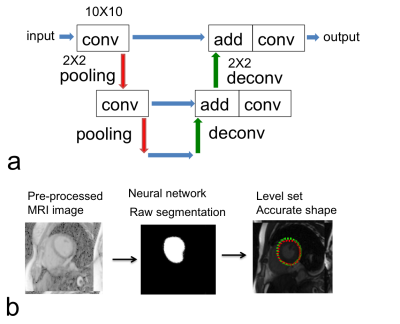
Image segmentation: (i) MRI images and LV labels were separated using random selection with 280-slice training and 32-slice testing datasets. Manual segmentation has been used as the ground truth to train the data and evaluate the automatic segmentation. (ii) The fully conventional network (FCN) was created based on an Unet structure [3] (Fig. 1a) with parameters: kernel size = 10×10, three decomposition levels, number of convolutional layers for the first/second level = 6, no convolutional layers for the third level, number of extracted features = 32, pooling size = 2×2, ReLU activation function and “add”-merging layers. For the training of both networks, the loss function was defined as negative logarithm of Dice coefficient. The data argumentation method includes shift (< 10%), rotation (< 20 degree) and zoom (< 20%), and raw MRI images were log-transformed before entering into the neural networks. All the neural networks were implemented in Tensorflow software (https://www.tensorflow.org/). (iii) The inferred shape used for initialization is also incorporated into elliptically refined level set model for segmentation. The new energy function of the proposed level set algorithm is: E=Elevel set+aEellipse+bEdeep learning. The proposed method combines the neural network and level set method (Fig.1b), where level set helps neural network with determining accurate elliptical shape of the myocardium.
Results and discussion
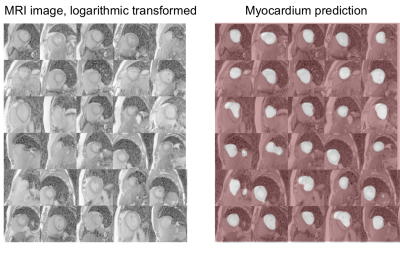
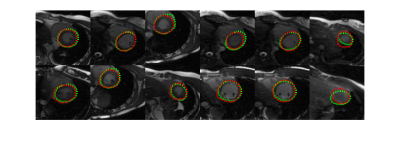
Fig.2 shows the representative results with neural network segmentation. Neural network was able to detect the location of the myocardium in most of the cases but could not determine the accurate elliptical shape of myocardium. Fig. 3 shows the final results from the combined use of neural network and level set method. The results obtained on 10 cases showed that the proposed segmentation was comparable with manual segmentation. And quantitatively, the average Dice coefficient of myocardium segmentation was measured as 76.22% and 81.78% using the local Gaussian distribution level set method [4] and FCN method respectively, and reached 84.27% using the proposed combined neural network and level set method.Conclusion
In summary, a novel method for fully automatic myocardium segmentation of 3D cardiac cine MRI has been developed. A larger clinical data cohort is needed for further testing the proposed method.Acknowledgements
This study is supported by grants NIH K25 EB014914, and NIH R56HL133663.References
[1] Liu J, Glenn OA, Xu D. Fast, free-breathing, in vivo fetal imaging using time-resolved 3D MRI technique: preliminary results. Quant Imaging Med Surg. 2014 Apr; 4(2):123-8. PMID: 24834424; PMCID: PMC4014872.
[2] Feng L et al., Highly accelerated real‐time cardiac cine MRI using k–t SPARSE‐SENSE. Magnetic Resonance in Medicine. 2013.
[3] Ronneberger O, Fischer P and Brox T. U-net: Convolutional networks for biomedical image segmentation In Proc. International Conference on Medical Image Computional and Computer-Assisted Intervention. 2015 234-241.
[4] Wang L, He L, Mishra A and Li C. Active contours driven by local Gaussian distribution fitting energy. Signal Processing. 2009 89 2435-47.
Figures