2727
Generalized AI for Organ Invariant Tissue Segmentation and Characterization of Multiparametric MRI: Preliminary ResultsVishwa Sanjay Parekh1, Katarzyna J Macura2,3, and Michael A Jacobs2,3
1Department of Computer Science, Johns Hopkins University, Baltimore, MD, United States, 2The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Sidney Kimmel Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
Artificial intelligence(AI) and deep learning techniques are increasingly being used in radiological applications. The true potential of deep learning in MRI applications can only be achieved by developing an AI that can learn the underlying MRI physics rather than a task that is specific to an organ or a particular tissue pathology. To that end, we developed and tested a multiparametric deep learning model capable of tissue segmentation and characterization in both breast cancer and stroke.
Introduction
Deep learning and other artificial intelligence(AI) techniques have begun to play an integral part in many different aspects of modern society1. Computer assisted clinical and radiological decision support have major impact from these technologies, with some excellent initial results2-10. However, the applications to date have been narrow, in that, they are restricted to a single task e.g. brain segmentation from multiparametric MRI(mpMRI). The true potential of AI will be achieved when the model is generalizable to multiple different tasks. One recent successful development of general AI came from Google DeepMind where they achieved human level performance in playing a large number of Atari games11. This study investigates the feasibility of a generalized deep network for segmentation and classification of mpMRI in different organs with contrasting tissue pathologies based on the underlying physical modeling of the MRI physics.Methods
We developed and validated a multiparametric deep learning(MPDL) breast MRI network for segmentation of breast lesions in 139 patients. Input into MPDL network were mpMRI of T1 and T2-weighted imaging, diffusion weighted imaging(DWI), apparent diffusion coefficient (ADC) mapping and dynamic contrast Enhanced(DCE) derived tissue signatures. Our segmentation MPDL network was constructed from stacked sparse autoencoders(SSAE) with five hidden layers12-14. The MPDL was trained and validated on the breast cancer dataset using k-fold cross validation with Dice Similarity(DS) metric. To apply the breast MPDL signatures to another mpMRI data set, we used stroke mpMRI of perfusion-, diffusion-, time-to-peak, T1-,T2-weighted imaging (PWI, DWI, TTP, T1WI and T2WI, respectively) obtained from five stroke patients at one, two or three time points(12 studies) after stroke: acute(≤12 hours), subacute(24-168 hours), and chronic(>168 hours). The phase resolution of PWI in test dataset was matched to the phase resolution of the training dataset using a combination of phase sliding window and wavelet decomposition15. The experiment was set-up as follows: Using the breast signature model, we applied the trained MPDL on the stroke dataset. Areas of infract and tissue at risk were segmented and tested against the MPDL results. The DS between the MPDL and Eigenfilter(EI) segmented stroke lesions was employed as the evaluation metric for validation of the MPDL segmentation stroke. Statistical significance was set at p≤0.05Results
The MPDL approach accurately defined and segmented both the breast and stroke data. Fig. 1 demonstrates the segmentation MPDL results on two malignant and two benign patients. The DS index between the lesion defined by EI and MPDL segmentation demonstrated excellent overlap with 0.87±0.05 for malignant patients and 0.85±0.07 for benign patients. More importantly, the results from MPDL tested on the independent stroke data were excellent as shown in fig. 2. There was high correlation between the quantitative ADC and TTP values corresponding to the lesion areas segmented by EI and MPDL (Rarea = 0.99, RTTP = 0.99, RADC=0.86) corresponding to infract and tissue at risk. Fig. 3 demonstrates the scatterplot and the Bland Altman plot between the quantitative values of ADC and TTP, and the lesion areas segmented by the EI and MPDL algorithms. Furthermore, the percentage difference in the lesion areas, TTP and ADC values segmented from EI and MPDL were 6±2%,3±3%, and 7±7% respectively.Discussion
The knowledge transfer between the MPDL trained on breast cancer and applied to stroke was excellent. These results demonstrate the robustness of the MPDL tissue signature model based on the underlying MR characteristics of each MRI parameter. For example, the T1 and T2 of the breast and stroke data were similar. The DWI and ADC mapping were the same, except for the number of b values. However, the MPDL method was able to adapt and apply the correct signatures. Deep learning has previously been successfully applied to segment and classify tissue types from brain, breast, prostate and other organs2-10. We successfully demonstrated the AI capability of deep learning algorithms to learn the underlying MRI physics. Our MPDL tissue signature model was able to segment different tissue types from pathologies (stroke) that the MPDL had never “seen” before.Conclusion
The MPDL model was able to accurately segment the tissue irrespective of the underlying organ or tissue pathology. Furthermore, the MPDL model reveals the possibility of universal deep learning model that can identify patterns in any organ or pathology or can be further personalized to different applications.Acknowledgements
Funding: R01CA190299, 5P30CA006973 (IRAT), U01CA140204, U01CA166104, U01CA151235 and equipment grant from Nvidia Corporation.References
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444.
- Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. May 2016;30:108-119.
- González AM. Segmentation of Brain MRI Structures with Deep Machine Learning. MS thesis. Universitat Politècnica de Catalunya, 2012.
- Guo Y, Gao Y, Shen D. Deformable MR Prostate Segmentation via Deep Feature Learning and Sparse Patch Matching. IEEE Trans Med Imaging. Apr 2016;35(4):1077-1089.
- Li R, Zhang W, Suk H-I, et al. Deep learning based imaging data completion for improved brain disease diagnosis. International Conference on Medical Image Computing and Computer-Assisted Intervention, 2014.
- Liu S, Liu S, Cai W, Pujol S, Kikinis R, Feng D. Early diagnosis of Alzheimer's Disease with deep learning. 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI)2014.
- Shin HC, Orton MR, Collins DJ, Doran SJ, Leach MO. Stacked Autoencoders for Unsupervised Feature Learning and Multiple Organ Detection in a Pilot Study Using 4D Patient Data. Ieee Transactions on Pattern Analysis and Machine Intelligence. Aug 2013;35(8):1930-1943.
- Suk HI, Lee SW, Shen D, Alzheimer's Disease Neuroimaging I. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain structure & function. Mar 2015;220(2):841-859.
- Xu J, Xiang L, Liu Q, et al. Stacked Sparse Autoencoder (SSAE) for Nuclei Detection on Breast Cancer Histopathology Images. IEEE Trans Med Imaging. Jan 2016;35(1):119-130.
- Zhang W, Li R, Deng H, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. Mar 2015;108:214-224.
- Mnih V, Kavukcuoglu K, Silver D, et al. Playing atari with deep reinforcement learning. arXiv preprint arXiv:1312.5602. 2013.
- Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. Advances in neural information processing systems. 2007;19:153.
- Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. Jul 2006;18(7):1527-1554.
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. Jul 28 2006;313(5786):504-507.
- Akhbardeh A, Jacobs MA, Methods and systems for registration of radiological images. US patent US90084622015.
Figures
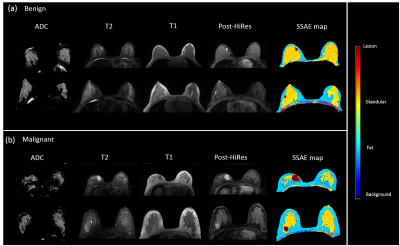
Figure 1. Illustrates the result
of MPDL network on axial breast mpMRI in (a) two representative benign patients
and (b) two representative malignant patients. The color coding is shown to the
right of the images.
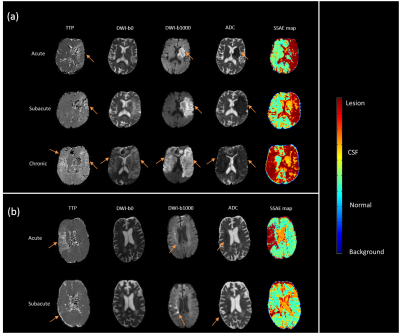
Figure 2. Illustrates the result
of MPDL network trained axial breast mpMRI and applied to stroke mpMRI in two
representative stroke patients at different time points after stroke. The color
coding is shown to the right of the images.
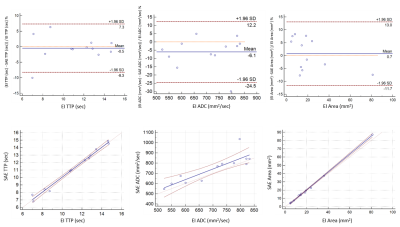
Figure 3. Bland Altman and scatterplots for comparison
between the Eigenfilter (EI) segmented stroke lesions and MPDL segmented stroke
lesions. (a) Time to peak (TTP). (b) Apparent diffusion coefficient map values. (c)
Lesion segmented areas.