2720
The Weakest Link in the Chain: How MR Data Quality influences Convolutional Neural Network Performance1Dept. of Radiology, Medical Physics, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany, 2Department of Radiation Oncology, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, German Cancer Consortium (DKTK), Partner Site Freiburg, Freiburg, Germany
Synopsis
In this work, tumor segmentation performance of a convolutional neural network is tested with respect to input data quality. 19 patients suffering from head and neck tumors underwent multi-parametric MRI including diffusion weighted imaging. The network was trained on multiparametric MR images with and without geometrically corrected diffusion data. With distortion correction, the Dice coefficient could be increased by 22% over uncorrected data showing the necessity for geometric image pre-processing in neural network analysis.
Introduction
Precise tumor segmentation is a crucial step in radiation therapy, as it decreases dose deposition in healthy tissue while maximizing treatment outcome. To assist physicians in the tedious task of segmentation, automatic detection methods have been proposed as tools for both diagnostics and treatment planning. With these automatic methods, current problems may be overcome such as inter-observer variability in target definitions, definition and assessment of tumor heterogeneity as well as tumor classification.
Many algorithms have been introduced for segmentation, starting from intensity-based local feature selection using clustering algorithms, atlas-based registration and segmentation, symmetry-based or feature learning using neural networks and even incorporating local and structural information with convolutional neural networks (CNN)1–9.
In this work a fully connected 3D CNN, DeepMedic2, is used. A plethora of studies analyzes the influence of network architecture on segmentation output – in this study, however, we investigate the effect of input data quality. Using data from a head and neck tumor patient study, the influence of image distortions in diffusion weighted imaging (DWI) on segmentation quality is quantified.
Materials and Methods
Patient Study
In total, 19 patients with large head and neck tumors have been included in the study. All patients underwent 3T MRI (Siemens Medical Solutions, Erlangen, Germany) with conventional Fast Spin Echo T1- and T2-weighted protocols, a dynamic contrast-enhanced (DCE) MRI for perfusion assessment (Ktrans maps), and multiecho GRE imaging for tumor oxygenation measurements (T2* maps). In addition, diffusion-weighted images (DWI) (b-values: 50, 400, 800 mm-2 s) were acquired to calculate tumor ADC maps.
Data Pre-Processing
Before training the CNN, T1- and T2-weighted images were normalized to zero mean and unary standard deviation. Ktrans, T2* and ADC are already in physically meaningful units so no further normalization was performed.
Head and neck regions are especially challenging for DWI as the complex geometry imposes severe limitations to magnetic field shimming10. To compare the CNN performance, both standard ADC maps and geometrically improved ADC maps were employed.
An algorithm was developed that estimates local geometric distortions between DWI and high-bandwidth T2-weighted images as ground truth. The resulting distortion field was used to correct the ADC maps.
Convolutional Neural Network
As input, gross tumor volumes (GTV) have been contoured by a radiation oncologist and a radiologist. These GTVs were used as ground truth labels for the network training and evaluation phase.
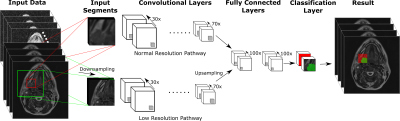
DeepMedic operated on multi-parametric 3D MRI data including T1- and T2-weighted images, Ktrans maps, T2* maps and ADC maps. Data was split to a high and a low resolution pathway to include both local and contextual information. Each pathway consisted of 8 hidden layers of {40 40 50 50 60 60 70 70} channels using 33 kernel sizes followed by 2 fully connected layers of 100 channels, which combine high and low resolution pathways (Figure 1).
Two separate neural networks were trained: the first network included original ADC maps while the second one was trained on geometrically undistorted ADC maps. The segmentation results of the 2 different networks without and with distortion correction was compared by calculating the Dice coefficients between the segmentation result for the tumor and the GTV.
Results
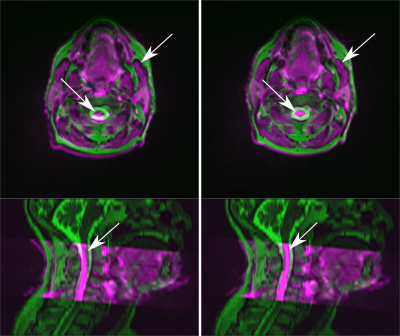
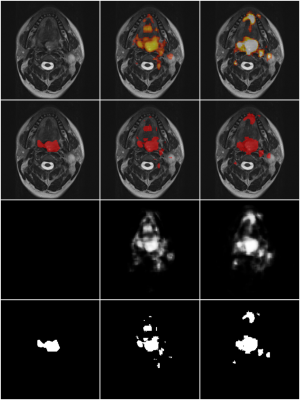
As expected, geometrical distortions in DWI are especially prominent in outer slices and closer to skin-air surfaces (Figure 2). Dice coefficients of the 2-class learning are 0.346 for the CNN without additional geometric correction, and 0.423 with correction. Exemplary segmentation results on patient data that was not included in the training process are shown in Figure 3. Note, that additional lymph node regions have been segmented although they were not labeled in the ground truth.Discussion
Correction of geometric distortions in DWI has a positive effect on segmentation performance of CNNs. The correction increased segmentation performance, even though Dice coefficients did not reach performances of similar networks2, which may be explained by the small set of only 19 patients. It shows that local image registration cannot be performed by the CNN itself, as its architecture does not allow for compensation of misregistrations. The proposed correction method is performed during preprocessing by connecting anatomical information of DWI and T2-weighted images without the use of a B0-fieldmap. Thus, segmentation results correspond better to the ground truth.Conclusion
Segmentation algorithms require high quality input data with known geometrical properties. This study in DWI data shows that distortion compensation is an essential pre-processing step in the data processing chain for CNNs which significantly improves segmentation outcome.Acknowledgements
This work was supported in parts by a grant from the Deutsche Forschungsgemeinschaft (DFG) under grant number HA 7006/1-1.References
1. Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004.
2. Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004.
3. Guo D, Fridriksson J, Fillmore P, Rorden C, Yu H, Zheng K, Wang S. Automated lesion detection on MRI scans using combined unsupervised and supervised methods. BMC Med. Imaging 2015;15:50. doi: 10.1186/s12880-015-0092-x.
4. Pinto A, Pereira S, Dinis H, Silva CA, Rasteiro DMLD. Random decision forests for automatic brain tumor segmentation on multi-modal MRI images. In: 2015 IEEE 4th Portuguese Meeting on Bioengineering (ENBENG). ; 2015. pp. 1–5. doi: 10.1109/ENBENG.2015.7088842.
5. Wilke M, de Haan B, Juenger H, Karnath H-O. Manual, semi-automated, and automated delineation of chronic brain lesions: A comparison of methods. NeuroImage 2011;56:2038–2046. doi: 10.1016/j.neuroimage.2011.04.014.
6. Schmidt P, Gaser C, Arsic M, et al. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. NeuroImage 2012;59:3774–3783. doi: 10.1016/j.neuroimage.2011.11.032.
7. Dupont C, Betrouni N, Reyns N, Vermandel M. On Image Segmentation Methods Applied to Glioblastoma: State of Art and New Trends. IRBM 2016;37:131–143. doi: 10.1016/j.irbm.2015.12.004.
8. Shah V, Turkbey B, Mani H, Pang Y, Pohida T, Merino MJ, Pinto PA, Choyke PL, Bernardo M. Decision support system for localizing prostate cancer based on multiparametric magnetic resonance imaging. Med. Phys. 2012;39:4093–4103. doi: 10.1118/1.4722753.
9. Prior FW, Fouke SJ, Benzinger T, et al. Predicting a multi-parametric probability map of active tumor extent using random forests. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). ; 2013. pp. 6478–6481. doi: 10.1109/EMBC.2013.6611038.
10. Oudeman J, Coolen BF, Mazzoli V, Maas M, Verhamme C, Brink WM, Webb AG, Strijkers GJ, Nederveen AJ. Diffusion-prepared neurography of the brachial plexus with a large field-of-view at 3T. J. Magn. Reson. Imaging 2016;43:644–654. doi: 10.1002/jmri.25025.
Figures