2719
Evaluation of 2D and 3D convolutional neural network methods for generating pelvic synthetic CT from T1-weighted MRI1David Geffen School of Medicine, UCLA, Los Angeles, CA, United States, 2Department of Radiation Oncology, UCLA, Los Angeles, CA, United States
Synopsis
Synthetic CT (sCT) must be generated directly from MRI scans to achieve MRI-only radiotherapy. We propose 2D and 3D convolution neural network models for generating pelvic sCT and evaluate their performance. Five-fold cross-validation is performed using paired T1-weighted MRI and CT scans from 20 patients. Our results show the 2D model generates accurate sCT for all patients in this study. The average mean absolute error (MAE) between CT and sCT across all patients is 38.0±3.9 HU in the 2D model. The average MAE is 55.9±28.4 HU in the 3D model. This large variation is possibly due to the limited number of 3D training volumes.
Introduction
Superior soft tissue contrast offered by magnetic resonance imaging (MRI) makes it a favored imaging modality for radiotherapy treatment planning.1-2 It is beneficial to use only MRI for planning, as standard practice involving both CT and MRI scans increases clinical workloads and raises the registration uncertainty3. A critical step to achieve MRI-only radiotherapy is to generate Hounsfield Unit (HU) maps, i.e. synthetic CT (sCT) images, from MRI scans. This is because MRI, unlike CT, cannot be used for dose calculation due to the lack of direct correspondence between MRI intensity and electron density. Han presented a SegNet-like 2D convolutional neural network (CNN) model which can produce accurate brain sCT.4-5 Here, we test CNN methods on pelvic anatomy. This is more challenging due to large inter-patient geometric variation. Our 2D CNN model is modified from SegNet5 and extended to 3D. The accuracy of pelvic sCT, produced by 2D and 3D CNN models, is then evaluated using voxel-wise and geometric metrics.Materials and methods
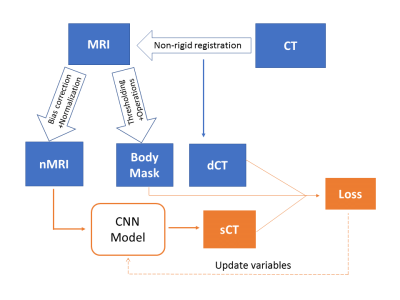
The overall workflow of sCT generation is shown in Figure 1. The T1-weighted MRI and CT scans of 20 prostate RT patients are retrospectively acquired. Thirty slices covering the pelvis are extracted from MRI scans and resampled to 256 × 256 ×30. The CT is registered to MRI using a multi-resolution B-spline algorithm in Elastix6. N4 bias filed correction7 and histogram-based normalization8 are performed on all MRI scans to ensure consistent MRI intensity throughout patients. A body mask of each patient, which is used to restrict loss evaluation and sCT accuracy assessment, is also generated from the MRI scan using Otsu's thresholding and morphological operations.
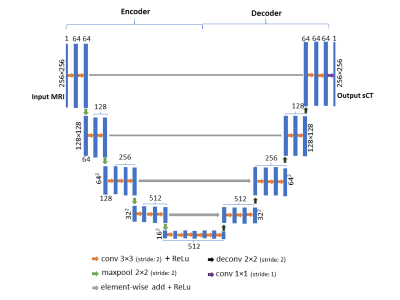
Our 2D and 3D CNN models are modified from SegNet5, a state-of-art deep learning architecture for semantic segmentation. First, de-convolution layers are used to replace upsampling layer to save computational memory. Second, residual shortcuts, connecting the encoder and decoder part, are added for faster convergence. Figure 2 shows the network architecture of the 2D model. The 3D model shares the same architecture except (1) all 2D operations are replaced with their 3D counterparts (2) a batch normalization layer is added after each 3×3×3 convolution layer for speeding up the training.
Both models are implemented using Tensorflow9 packages. On-the-fly data augmentation, such as random shift and rotation, is performed to reduce overfitting. The loss function is defined as mean absolute error (MAE) between sCT and deformed CT (dCT) within the body mask plus an L2 regularization term, i.e. $$loss=\frac{1}{N} \sum_{i=1}^{n}||sCT(i)-dCT(i)||+\lambda\sum_{i=1}^{k}ω_i^2$$
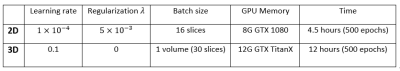
which is minimized using Adam stochastic gradient descent method with default parameters except for initial learning rate.10 Since the convolution layers of the VGG16 network11 is identical to the encoder part of the 2D model, we initialize the weights of 2D encoder part using pre-trained VGG16 weights. All other weights of convolution filters are initialized using Xavier initialization12, and all bias parameters are initialized to 0. The details of training for two models are summarized in Figure 3.
Results
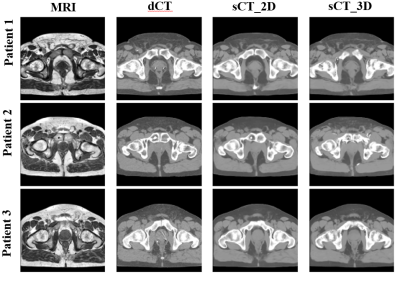
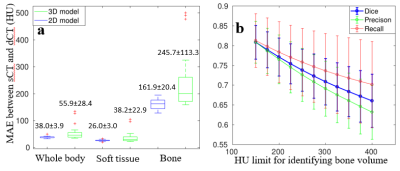
The sCT of every patient is generated in five-fold cross-validation framework. It only takes about 4 seconds to produce a sCT volume. Figure 4 shows three patients' sCT images with corresponding MRI-dCT image pairs. The 2D and 3D model generates sCT images which look similar to dCT images. However, the 3D model is not robust as shown in Figure 5a, the box-whisker plot of the MAE for different tissues across all patients. Cortical bone is mapped to soft tissue in 3D sCT for three patients as indicated in red cross above bone boxes of the 3D model.
Besides voxel-wise accuracy, as shown in Figure 5b we also present the dice similarity coefficient ($$$ \frac{TP}{2TP+FP+FN} $$$), recall ($$$ \frac{TP}{TP+FN} $$$) and precision( $$$ \frac{TP}{TP+FP} $$$) of bone volumes, which are defined using different HU thresholds, between dCT and sCT generated in the 2D model.
Discussion and conclusion
We present 2D and 3D CNN models for generating pelvic sCT using T1-weighted MRI. In our study, the 2D model successfully generates accurate sCT for all patients with average MAE at 38.0±3.9 HU. Although the 3D CNN model is supposed to catch more information like global geometry, its performance is not robust. This may be caused by limited number of 3D volumes available for training. The fast generation speed and accurate HU mapping make proposed models, especially the 2D model, promising tools to generate pelvic sCT for future MRI-only radiotherapy. More patient data is required for improving the performance of the 3D model, and dose comparisons between CT and sCT are required for clinical implementation.Acknowledgements
No acknowledgement found.References
[1] Rasch, C., Steenbakkers, R., & van Herk, M. (2005, July). Target definition in prostate, head, and neck. In Seminars in radiation oncology (Vol. 15, No. 3, pp. 136-145). WB Saunders.[2] Eskander, R. N., Scanderbeg, D., Saenz, C. C., Brown, M., & Yashar, C. (2010). Comparison of computed tomography and magnetic resonance imaging in cervical cancer brachytherapy target and normal tissue contouring. International Journal of Gynecological Cancer, 20(1), 47-53.[3] Roberson, P. L., McLaughlin, P. W., Narayana, V., Troyer, S., Hixson, G. V., & Kessler, M. L. (2005). Use and uncertainties of mutual information for computed tomography/magnetic resonance (CT/MR) registration post permanent implant of the prostate. Medical physics, 32(2), 473-482.[4] Han, X. (2017). MR‐based synthetic CT generation using a deep convolutional neural network method. Medical Physics, 44(4), 1408-1419.[5] Badrinarayanan, V., Kendall, A., & Cipolla, R. (2015). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv preprint arXiv:1511.00561. [6] Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. (2010). Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging, 29(1), 196-205.[7] Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan, A., Yushkevich, P. A., & Gee, J. C. (2010). N4ITK: improved N3 bias correction. IEEE transactions on medical imaging, 29(6), 1310-1320.[8] Nyúl, L. G., Udupa, J. K., & Zhang, X. (2000). New variants of a method of MRI scale standardization. IEEE transactions on medical imaging, 19(2), 143-150.c[9] Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z., Citro, C., ... & Ghemawat, S. (2016). Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467.[10] Kingma, D., & Ba, J. (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.[11] Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.[12] Glorot, X., & Bengio, Y. (2010, March). Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics (pp. 249-256).Figures