2718
Noninvasive Identification of IDH-mutational Status from 1H-MRS Spectra by Deep Learning1Department of Biomedical Sciences, Seoul National University, Seoul, Republic of Korea, 2Department of Radiology, Seoul National University Hospital, Seoul, Republic of Korea
Synopsis
Noninvasive identification of IDH-mutational status in glioma patients using 1H-MRS is diagnostically and prognostically valuable. However, the most widely used short TE method is reported to be more subject to false diagnosis due to the severe spectral overlap of 2HG. We explored the potential applicability of deep learning in addressing this issue. A deep neural network that was trained on a large number of simulated spectra substantially improved the overall diagnostic accuracy on the patient spectra, compared to the LCModel analysis. As no spectral fitting is involved, our results are not subject to ambiguity arising from the CRLB-based data interpretation.
Target audience
Those interested in noninvasive identification of IDH-mutational status in glioma patientsIntroduction
Noninvasive identification of isocitrate dehydrogenase (IDH)-mutational status in glioma patients using 1H-MRS is diagnostically and prognostically valuable, which targets detection of the onco-metabolite, 2-hydroxyglutarate (2HG).1-3 Due to severe spectral overlap of 2HG, spectral editing may be advantageous over the short TE method (data acquisition at short TE followed by spectral fitting), which may be more subject to false positive/negative diagnosis.1,4 However, the short TE method is still the most widely used method with high SNR and no need for sequence modification. Furthermore, the performance of the short TE method can be improved by incorporating those metabolites that are also altered in the IDH-mutated gliomas5,6 into the diagnostic criteria.6 Given the widely spread spectral distribution of those IDH-mutation-associated metabolites, therefore, there may be a spectral feature that can be captured by deep neural networks (DNNs) for differentiation between spectra with and without IDH-mutation.
We explored potential applicability of DNNs in noninvasive identification of IDH-mutational status in glioma patients. A DNN was trained and tested on simulated spectra and then evaluated on patient spectra.
Methods
Patient data : The study was approved by IRB (13 brain tumor patients). Spectra were collected from the brain tumor of the patients using PRESS at 3.0T (Siemens; TR/TE=2000/30 ms, SW=2 kHz, 2048 points, 64-128 averages, voxel size=8-17 cc). Patient-specific metabolite-nulled spectra were also acquired as a surrogate of spectral baseline (IR-PRESS; TI=680 ms). Following MRS scan, patients underwent surgery, and the collected tissues were sent for gene sequencing. The patient spectra were analyzed by LCModel for comparison.
Simulation of spectra for training and test of DNN : Before simulation, a spectral basis set was obtained for 18 metabolites from chemical phantoms. To simulate spectra of brain tumor, baseline-subtracted patient spectra were overviewed and the 1.1-4.5 ppm range was divided into 12 regions. In each region except the 1.7-2.4 ppm region where the MNPQ multiplet of 2HG (AMNPQ spin-system) resides, the relative signal intensity ratio of the metabolites resonating in that spectral region was determined by maximizing the correlation and minimizing the mean-squared-error between the simulated spectrum and patient spectrum for each patient. Then, the mean value over all patient spectra was used as the relative signal intensity ratio for Ala, Asp, Cr, Glc, GPC, Lac, mI, PCh, PCr, PE, and Tau. For the 1.7-2.4 ppm region, the relative ratios of 2HG, GABA, Glu, Gln, GSH, NAA, NAAG were varied individually from 0 to 5, 10, or 15 (step size=1). This generated 228,096 spectra without 2HG and 2,280,960 spectra with the relative ratio of 2HG varying from 1 to 10. From each of these two spectra groups with and without 2HG, 50,000 spectra were randomly selected, and line-broadened with 15 different linewidths for each spectrum followed by SNR adjustment to mimic in vivo spectra. This resulted in 750,000 spectra for each of the two spectra groups. Finally, 600,000 spectra were randomly selected from each of the 2HG-present and 2HG-absent groups and used as a training data set (n=1,200,000). The rest 300,000 spectra were used as a test data set.
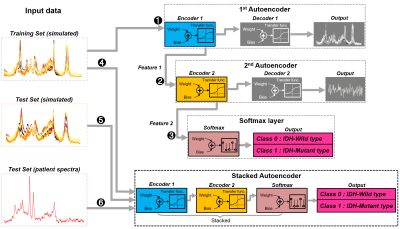
DNN : A stacked autoencoder7 was designed (Matlab, Mathworks Inc.) and trained on the simulated spectra for binary classification of the spectra into either IDH-mutant or wild-type (Figure 1). The trained DNN was tested first on the simulated spectra and then on the baseline-subtracted patient spectra.
Results
There were 12 glioma and 1 lymphoma patients. Among the glioma patients, there were 7 IDH-mutant and 5 wild-type patients. One lymphoma patient was classified as wild-type. The LCModel analysis of the patient spectra obtained a sensitivity of 14% (1/7) and a specificity of 83% (5/6) with an overall diagnostic accuracy of 46% (6/13). The DNN obtained ~100% accuracy on the simulated test set, and resulted in a sensitivity of 71% (5/7) and a specificity of 83% (5/6) with an overall diagnostic accuracy of 77% (10/13) on the patient spectra.Disccusion
To reduce the amount of simulation, no baseline was considered in the simulated spectra and thus the DNN was tested on the baseline-subtracted patient spectra. The metabolite concentrations were varied only for 7 metabolites in the simulation. Nonetheless, the overall diagnostic accuracy was substantially improved (from 46% to 77%) by the DNN. As no spectral fitting is involved, our DNN-based spectra classification is not subject to bias and ambiguity arising from the use of Cramér-Rao-lower-bounds (CRLB) as a measure of fitting reliability8 (e.g., 2HG fitted with CRLB>20%).Conclusion
Our preliminary results support potential applicability of deep learning in noninvasive identification of IDH-mutational status in glioma patients.Acknowledgements
This work was supported by the Ministry for Health, Welfare & Family Affairs (HI13C0015) and the NRF funded by the Ministry of Education, Science and Technology (2016R1D1A1B03931233) of Korea.References
1. Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116): 116ra4-116ra4.
2. Choi C, et al. 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nature. 2012;18:624-629.
3. Pope WB, et al. Non-invasive detection of 2-hydroxyglutarte and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197-205.
4. Choi C, et al. A comparative study of short- and long-TE 1H-MRS at 3T for in-vivo detection of 2-Hydroxyglutarate in brain tumors. NMR Biomed. 2013;26(10):1242-1250.
5. Kim H, et al. In-vivo proton magnetic resonance spectroscopy of 2-hydroxyglutarate in isocitrate dehydrogenase-mutated gliomas: A technical review for euroradiologists. Korean J Radiol. 2016; 17(5):620-632
6. Nagashima H, et al. Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro Oncol. 2016;18(11): 1559-1568
7. Bengio Y, et al. Greedy layer-wise training of deep networks. In: Advances in neural information processing systems. 2007;153-160.
8. Kreis R. The trouble with quality filtering based on relative Cramér-Rao lower bounds. Magn Reson Med. 2016;75(1):15-18.
Figures