2716
A multi-channel convolutional neural network for segmentation of breast lesions in DCE-MRI1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 3Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Radiomics offers a highly quantitative and high-dimensional view of the tumor microenvironment which no conventional imaging technique allows. It is the ideal strategy for personalizing care in heterogeneous cancers such as in the breast. Most approaches require time consuming, manual region of interest segmentation. Here, we present a fast and accurate neural network approach for breast lesion segmentation which can be adapted to accept any number of imaging modalities and shows reliability across many types of lesion.
Introduction
Radiomic analysis provides high-dimensional, multifaceted data about tumor microenvironment and physiology which are not apparent on standard medical images1. The ability to extract such a wealth of information from common-practice radiological studies would significantly improve the standard of care for highly heterogeneous diseases such as breast cancer.
Despite the exciting avenues radiomics provides, it has found little clinical adaptation. This is largely because lesion volumes often need to be manually drawn by radiologists, adding a significant amount of time and work required for an image to be analyzed. Recently, convolutional neural networks (CNN) have become recognized as powerful tools for a variety of image processing and computer vision tasks. Here, we apply a highly adaptable CNN, capable of integrating data from essentially any MRI pulse sequence or other diagnostic image, to the task of breast tumor segmentation.
Methods
Previously acquired clinical DCE-MRI data from 316 women (age: 57±12 years) with histopathology-confirmed breast cancer were extracted for this analysis with approved IRB exemption. Table 1 lists the different types of lesions included, and their frequency. Manual lesion segmentation was performed by an experienced breast radiologist, these manually drawn segmentations were used as the ground truth for CNN evaluation. Data were randomly partitioned into training (n=244), testing (n=36) and validation (n=36) sets.
The CNN architecture employed is shown in Figure 1. We trained a U-shaped CNN2 for direct end-to-end generation of tumor segmentation maps. All 2D convolutional layers employed zero-padding of their input and a stride of one so as not to change the size of the derived features. The network consists of a series of horizontal levels with increasing numbers of derived features being computed on increasingly downsampled representations of the input. Levels are connected by maxpooling operations of stride 2, which afford efficient downsampling, and stride 2 transposed convolutions, which upsample derived features for further computation in higher levels. L2-regularizaiton was used to prevent overfitting. A sigmoid function was used to convert voxel-wise outputs to probabilities ranging between 0 and 1. Loss was defined as the cross entropy between output probability maps and ground truth images. All training and testing were performed with Tensorflow. The network was trained for 50 epochs. CNN input consisted of the first two volumes in the DCE series (pre-contrast and wash-in phases). Data were fed in as successive sagittal slices, with corresponding pre-contrast and wash-in slices grouped as channels of a single input. Segmentations were generated by hard thresholding of lesion probability maps at 0.3. Binarized segmentations were compared to the radiologist’s manual segmentations. Reported metrics are the Dice coefficient and sensitivity.
Results
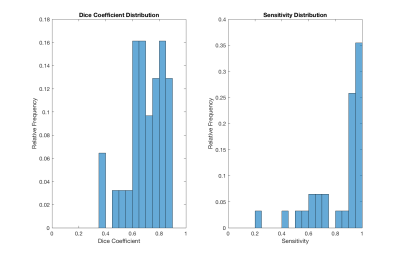
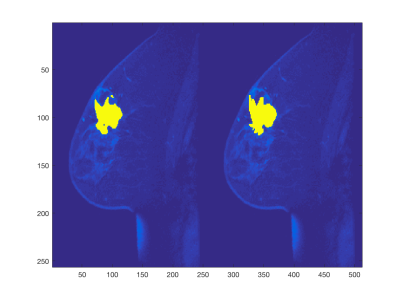
The training phase completed 50 epochs in 8.6 hours on a server equipped with Tesla K80. In the independent testing set, we observed a mean sensitivity of 0.83 (±0.19) and a mean Dice coefficient of 0.70 (±0.13). Histograms of these performance metrics are shown in Figure 2. An example segmentation for an irregularly shaped lesion is shown in Figure 3.Discussion
Automated breast lesion segmentation has been of interest for many years. Here, we have demonstrated the applicability of the U-shaped CNN to this end.
The neural network we employed achieved Dice coefficients similar to other recently published techniques for segmenting lesions out of DCE-MRI volumes3,4. The approach here substantially improves upon the state of the art in several noteworthy ways. CNN-based segmentation is rapid following initial training; this highly amortized cost allows for efficient application to both clinical and research pipelines, new MRI images can be segmented in a matter of seconds even using a CPU. Secondly, the network is almost entirely convolutional, placing only modest constraints on the size and shape of input images. This flexibility is also exhibited by our multi-channel design, which can incorporate an arbitrarily large amount of multimodal inputs; the network is in no way reliant upon underlying assumptions regarding the presentation of lesions in the input domain.
Methodologically, our approach is likely more translatable to broad clinical practice. We utilized a large population with considerable diagnostic heterogeneity, directly reflecting the population presenting for breast cancer treatment at our medical center, and present a technique which can seamlessly incorporate any number of imaging modalities. This is in contrast to recently published breast segmentation studies which, although achieving similar accuracy, limited their study sample to a single breast cancer subtype3, or employ methods which are highly dependent upon a specific modality4.
Conclusion
U-shaped CNNs provide a flexible and capable tool for segmenting lesions in breast DCE-MRI images.Acknowledgements
No acknowledgement found.References
1. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278(2):563-577.
2. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention2015.
3. Jayender J, Chikarmane S, Jolesz FA, Gombos E. Automatic segmentation of invasive breast carcinomas from dynamic contrast‐enhanced MRI using time series analysis. Journal of Magnetic Resonance Imaging. 2014;40(2):467-475.
4. Maicas G, Carneiro G, Bradley AP. Globally optimal breast mass segmentation from DCE-MRI using deep semantic segmentation as shape prior. Paper presented at: Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on2017.
Figures