2681
Influence of Parameter Optimization and Segmentation on the Accuracy of Various Registration Approaches for Multi-parametric 3D Breast MRI Data1Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India, 2C-DOT India, New Delhi, India, 3Computer Science and Engineering, Indian Institute of Technology Delhi, New Delhi, India, 4Department of Radiology, Fortis Memorial Research Institute, Gurgaon, India, New Delh, India, 5Department of Biomedical Engineering, All India Institute of Medical Sciences Delhi, New Delhi, India
Synopsis
Registration of human Breast MRI images is challenging due to its elastic deformable nature. In this study, various existing rigid and non-rigid registration methods were evaluated and compared in terms of accuracy and computation time. This work investigated influence of different registration parameters and showed possible ways to achieve better registration results. Experiential result revealed that the combination of Affine and B-spline method provided more time efficiency and accuracy than other methods.
Introduction:
Multi-parametric Breast MRI images can be used to generate quantitative parameters which enable an improved Breast tumor diagnosis, staging with high sensitivity and specificity.(1-3). Quantitative analysis requires T1-map(computed using PD-W,T1-W and T2-W images), multiple pre and post contrast gadolinium images. Any motion between MRI sequences due to breathing, patient positioning etc. can affect the accuracy of semi-quantitative and quantitative parameters which may mislead the clinician. Therefore, 3D-registration of Breast MRI images is an essential prerequisite to mitigate motion related artifacts before analysis. Registration of Breast MRI images is challenging due to absence of standard template and a large variation in shapes and sizes of the Breast4. The objective of this study was to evaluate and compare various existing rigid and non-rigid registration(NRR)(2,4) methods in terms of accuracy and computation time. This study also investigated the impact of segmentation and the influence of parameter tuning in registration. Registration results were assessed using validation methods(VM) such as Dice-Coefficient, Hausdroff-Distance and based on perfusion parameters(3,6).Methods:
All MRI experiments of were performed at 3T-whole body Ingenia MRI system(Philips-Healthcare, The Netherlands) using a 7-channel biopsy compatible breast coil. In this study, we have included twenty female subjects having Breast tumor.
MRI Data acquisition: After a localizer, T1, T2 and PD weighted(W) images, with and without fat saturation were acquired using turbo-spin-echo pulse sequence. Fat saturation was based upon DIXON method8. Multiple slices, covering entire breast tissue with slice thickness=3mm were acquired for all three data types. FOV=338×338mm2 and matrix size=452×338(interpolated matrix=512×512) were used. For PD weighted, TR/TE=2974ms/30ms was used. For T2-W, TR/TE=2974ms/100ms was used. For T1-W, TR/TE=603ms/10ms was used. T1-perfusion MRI was performed using a 3-dimensional-fast-field-echo(3D-FFE) sequence(TR/TE=3.0ms/1.5ms, flip-angle=12o, FOV=338×338mm2, slice thickness=3mm, matrix size=228×226(interpolated matrix=512×512) and acquisition time=3.7minute).
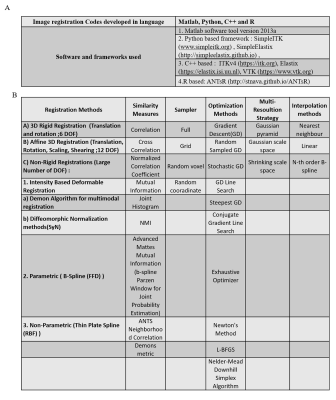
Data processing: Registration codes were developed using multiple registration frameworks(Figure-1[A]). These codes were executed on a system with 8-core Intel Xenon CPU@3.2GHz processors with 16GB-RAM. The first time-point(pre-contrast) in 3D-image stack of T1-perfusion MRI sequence, considered as reference image, was used to register structural image(T1-W, T2-W, PD-W) and subsequent 3D-image stacks of T1-perfusion MRI at different time point. Various 3D-registration methods were evaluated on breast MRI images(with and without segmentation) combining different similarity measures and optimization algorithms from Figure-1[B]. The performance of various registration methods was evaluated using validation methods such as Dice-Coefficient, Hausdroff-Distance and based on quantitative parameters. For further validation, SI-curve of voxels near boundary of tumor were compared with SI-curves of voxels in the center of tumor (homogeneous region) using Affine, B-spline with Affine, Symmetric-image-Normalization7(SyN) registration approaches. Data were analysed using generalized-tracer-kinetic model for computing Ktrans maps before and after registration.
Results:
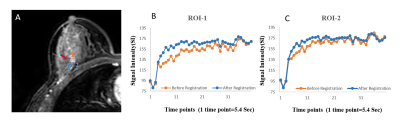
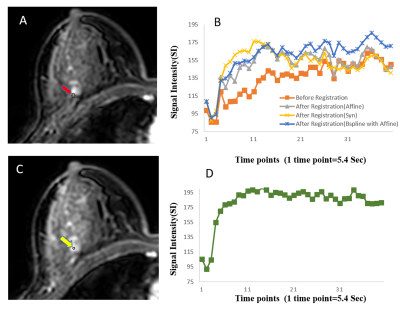
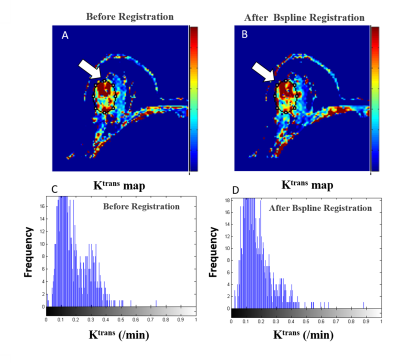
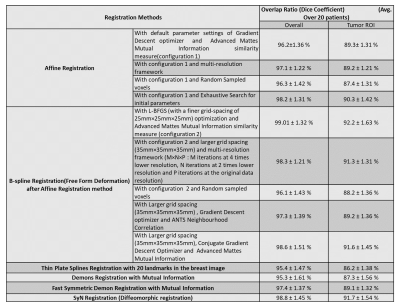
Registration results are shown in Table-I. B-spline and SyN methods showed higher overlapping ratio than others. The improvement of SI-curve before and after registration within tumor ROIs highlight the need of registration(Figure-2). Among all these methods, B-spline with Affine provided higher correlation between SI-curve from boundary voxels and center(homogeneous) region of the tumor(Figure-3). In Figure-4, histogram after registration(B-spline with Affine) was smoother as compared to before registration. Similarly, the improvement of Ktrans value after registration signifies the need of motion correction before quantitative analysis. The accuracy and time were significantly improved using segmented Breast images. The registration of a segmented Breast data containing T1-W, T2-W, PD-W(each data has 60-slices) and DCE-MRI data(60-slices with 40 time-points) took 60-80 minutes using B-spline NRR with limited-memory BFGS(L-BFGS) optimizer(With grid-spacing=35mm×35mm×35mm and multi-resolution=4×2×1). It can be reduced to 10-12 minutes using parallel-processing(tested with parfor in Matlab2013a).Discussion:
B-spline with Affine and SyN methods were better than other methods at correctly aligning breast MRI images, whereas demon methods failed to provide consistent results. Among all these methods, B-spline with Affine provided accurate results in this study. In case of without-segmentation, registration took more time and evaluation results degraded. For the initial-guess of optimizer parameters, methods like exhaustive, random search in a range of values, evolutionary algorithm can be used to have optimal registration. Parallel-processing can be useful in searching good initial point to save time. Registration processing time depends on voxel sampling percentage, iteration-number, grid-spacing etc. The choice of optimizer, tuning of optimizer-parameters, choice of metric, histogram-bins in metric etc. played crucial role in registration. These parameters can be tuned for optimizing registration time and improving the accuracy.Conclusions:
The combination of Affine and B-spline method provided more time efficiency and accuracy than other methods in this study. B-spline NRR may introduce new deformations and higher memory usage with very low grid spacing, whereas SyN has achieved consistent performances with no new deformations introduced but sometimes it may be time consuming.Acknowledgements
The authors acknowledge an internal funding support from IIT-Delhi. Authors acknowledge support of Philips India Limited and Fortis Memorial Research Institute Gurugram in MRI data acquisition. Authors thank Prof. Rahul Garg for providing technical support, Dr.Vedant Kabra for clinical input and Dr Pradeep Gupta for data handling.References
1. Jennifer A. Harvey et. al. Breast MR Imaging Artifacts: How to Recognize and Fix Them (2007).
2. D. Rueckert et. al. Non-rigid Registration of breast MR images using Mutual Information (2006).
3. Melbourne A et. al. The effect of motion correction on pharmacokinetic parameter estimation in dynamic-contrast-enhanced MRI(2011).
4. D. Rueckert et. al. Nonrigid registration using free-form deformations: application to breast MR images(1998).
5. Yago Diez et al. Comparison of Methods for Current-to-Prior Registration of Breast DCE-MRI(2014).
6. Andrew Hill et al. Evaluating the accuracy and impact of registration in dynamic contrast-enhanced breast MRI(2009).
7. Avants BB et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labelling of elderly and neurodegenerative brain (2008).
8. Dixon W. Simple proton spectroscopic imaging. Radiology (1984).
Figures