2660
Reduction of respiratory motion artifact in c-spine imaging using deep learning: Is substitution of navigator possible?1Yonsei University, Seoul, Republic of Korea, 2Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
Synopsis
Deep learning methods are starting to be widely used in medical images. Here, we propose a deep learning approach to compensate respiratory induced artifacts. A deep convolutional neural network was designed to train the ghosting artifact caused by respiratory motion in c-spine imaging. Using deep learning, compensation can be applied without additional data such as navigator echo.
Introduction
In c-spine imaging, GRE based imaging is typically performed to distinguish hemorrhage, calcification, etc1. GRE based methods are inherently susceptible to spatial and temporal B0 variations such as respiration. Respiratory motion is more evident in cervical spinal cord because of the proximity of the respiratory-induced organs such as lung. Respiration makes temporal B0 fluctuations, generating artifacts such as ghosting especially in the phase encoding direction. To overcome these artifacts, navigator methods have previously been presented2. While previous approaches using deep learning for motion correction used fabricated images for training3, here, the labeled data set comes real data sets with navigator correction. Hence, we propose a deep learning approach to compensate respiratory induced artifacts as a potential method for replacement of navigators.Methods
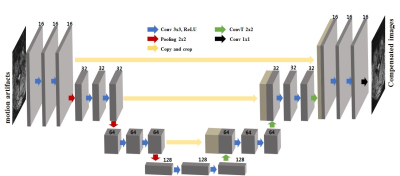
The U-net architecture was used for deep neural network which has gained high reputation for medical imaging purposes4. The structure includes convolution, batch normalization, rectified linear unit, contracting path connection, max pooling, and convolution transpose (Fig 1).
Data were acquired using 2D multi-echo GRE on a clinical 3T MRI scanner (Tim Trio, Siemens Medical Solution) with the following parameters: TR=968 ms, TE=20ms, FA=68, 16~20 slices per subject, 0.5x0.5 mm2 in-plain resolution with 3 mm slice thickness. matrix size is 384x384 and GRAPPA (factor 2) was applied. A navigator echo time was placed at 7 ms.
A total of 170 slices of MR images from 9 healthy volunteers were used for the training and testing. To overcome lack of data, data augmentation was performed by left/right reversal of the training samples. Overall, 300 slices from 8 subjects were used for training, and 20 slices from another subject were used for testing. The input data were corrupted images from respiratory motion. The label data were compensated images using the navigator. Therefore, we are comparing our deep learning method with navigator corrected images since obtaining motion-free data in this region is difficult.
Results and Discussion
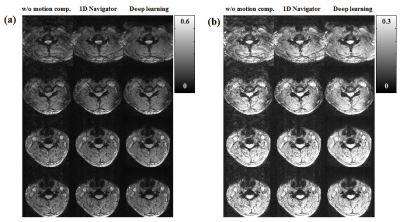
Fig. 2 shows representative slices of the input images (motion corrupted images), output images (compensated images using deep learning approach), and labeled images (compensated images using 1D navigator). Ghosting and cloudy-like artifacts caused by respiratory motion are reduced using both 1D navigator and deep learning approach.
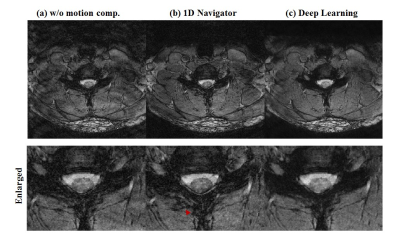
Fig. 3 shows another representative example. Since the network used in this study used navigator compensated images as a label, the results of learning should follow the results of 1D navigator. However, in the compensated image with 1D navigator, unwanted image distortion is observed (Fig. 3 red arrow) which is not noticed in the results of deep learning. This is because 1D navigators are used to compensate for rigid motion and non-rigid motion can cause errors. This error might be reduced when well-trained networks are used. In addition, when the network is built using data compensated for non-rigid motion, better quality results could be obtained using the deep-learning approach. Compensated images with deep learning approach shows slightly blur in some regions which possibly can be improved with more training.
Conclusion
We proposed a respiratory artifact compensation method using a deep convolutional neural network. In this study, compensation was performed based only on the c-spine magnitude images from real scans. Further study is needed to determine whether phase images can be compensated. It is seen that deep learning based correction can potentially substitute navigators. The method can be expanded to other regions where navigators are commonly used.Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Science, ICT and future Planning (NRF-2016R1A2B3016273)References
1. Thomas B, Somasundaram S, Thamburaj K, Kesavadas C, Gupta AK, Bodhey NK, Kapilamoorthy TR. Clinical applications of susceptibility weighted MR imaging of the brain–a pictorial review. Neuroradiology 2008;50(2):105-116.
2. Wen J, Cross AH, Yablonskiy DA. On the role of physiological fluctuations in quantitative gradient echo MRI: implications for GEPCI, QSM, and SWI. Magnetic resonance in medicine 2015;73(1):195-203.
3. SCHÖLKOPF, Bernhard. Retrospective Motion Correction of Magnitude-Input MR Images. In: Machine Learning Meets Medical Imaging: First International Workshop, MLMMI 2015, Held in Conjunction with ICML 2015, Lille, France, July 11, 2015, Revised Selected Papers. Springer, 2015. p. 3.
4. O. Ronneberger, P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. In MICCAI. 2015. P. 234–241
Figures