2560
Performance of PIRADSv2 ≥3 and ≥4 scores as cut-offs for the detection of prostate cancer1Department of Radiodiagnosis, All India Institute of Medical Sciences, New Delhi, India, 2Department of Urology, All India Institute of Medical Sciences, New Delhi, India, 3Department of NMR, All India Institute of Medical Sciences, New Delhi, India
Synopsis
We report a prospective evaluation of prostate imaging reporting, archiving and data system version 2 (PIRADSv2) for multiparametric MRI (mpMRI) taking histopathology of radical prostatectomy specimens as
Introduction
Prostate imaging reporting, archiving and data system version 2 (PIRADSv2) is the currently recommended standardized reporting for lesion appearing on multiparametric MRI (mpMRI) for detection of cancer of prostate (CaP). Most of the studies evaluating PIRADSv2 used systematic or targeted biopsies as reference standards and all of them are retrospective studies making use of data before the publication of PIRADSv2. Therefore, in the present study, we prospectively evaluated the performance of PIRADSv2 taking radical prostatectomy specimens as the reference standard and determined the optimal cut-off for scores to detect clinically significant CaP.Methods
26 patients (mean age, 63.3 ± 7 years; mean PSA, 12 ± 5 ng/ml) having a biopsy-proven CaP, were investigated using mpMRI, followed by radical prostatectomy within 1 month. mpMRI was carried out at 3.0T MRI scanner (Ingenia, Philips) using a phased array flex coil. mpMRI protocol consisted of T2W, T1W imaging, DWI (single-shot echo-planar imaging sequences; b-values 0, 500, 1000 and 1500 s/mm2) and DCE-MRI. T1W spoiled gradient-echo sequence, with Gadodiamide, 0.1 mmol/kg, at a rate of 3ml/sec was used for DCE-MRI. The radical prostatectomy specimens were sectioned into 3mm cuts, in the axial plane to determine the Gleason grade group. Prostate images were divided into 16 sectors (12, peripheral zone (PZ) and 4, transitional zone (TZ) sectors) per patient. Each of these sectors was assigned a PIRADSv2 score for T2WI, DWI and DCE-MRI individually and a score for the sector. Cancer positive sectors which showed PIRADSv2 ≥ 3 lesions were then divided into 3 groups; (i) Low-grade cancer (LGC), sectors with a histopathologically detected tumour with Gleason grade group 1, with a corresponding lesion on mpMRI, (ii) Intermediate-grade cancer (IGC), sectors with a histopathologically detected tumour with Gleason grade group 2 or 3, with a corresponding lesion on mpMRI. (iii) High-grade cancer (HGC), sectors with a histopathologically detected tumor with Gleason grade group 4 or 5, with a corresponding lesion on mpMRI.Results
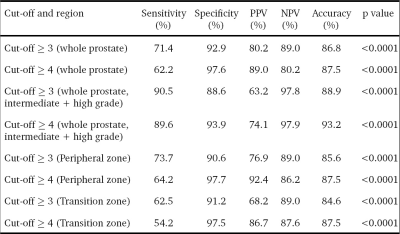
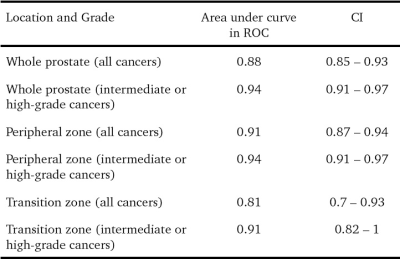
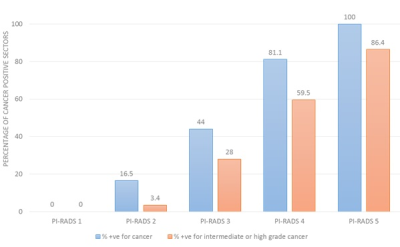
Out of the 416 sectors which were assessed [312 PZ + 104 TZ], 119 (28.6%) were positive for cancer on histopathology. 95 of these 119 positive sectors (79.8%) were from the PZ while 24 (20.2%) of them were from the TZ. The median number of cancer-positive sectors per prostate was 2.5 (range 1-16). A cut-off ≥ 4 score showed a slightly higher accuracy than a cut-off ≥ 3 (87.5% vs 86.8%), however, sensitivity (62.2% vs 71.4%) and NPV (80.2% vs 89%) were reduced (Table 1). All high-grade cancers were correctly classified as cancer, irrespective of the cut-off used. Among the cancers not detected at ≥4, 45.2% (n=14/31) were intermediate-grade, while among those not detected at ≥3, only 25.9% (n=7/27) were of the intermediate grade. When assessed for the group of intermediate and high-grade cancers, a cut-off of ≥ 4 showed the best specificity (93.9%) and accuracy (93.2%) without compromising on sensitivity (89.6%). The prostate cancer positivity rates for each of the PIRADSv2 assessment categories are presented in Fig. 1. 16.5% of PIRADSv2 score 2 lesions turned out to be cancer on histopathology, most of these were low-grade cancers. The discriminative abilities of PIRADSv2 using ROC curve are summarised in Table 2.Discussion
We assessed the accuracy of PIRADSv2 for the detection of CaP and determined ≥ 3 score to be an optimal cut-off. There is a wide variation in sensitivity, specificity, positive and negative predictive values reported for the performance of PIRADSv2. The possible contributing factors to wide variation in sensitivity and specificity may be due to different cut-offs, acquisition protocols, study designs and subjectivity introduced by users into PIRADSv2 reporting. To our knowledge, this is the first study to prospectively evaluate PIRADSv2, strictly adhering to their imaging acquisition protocol recommendations. High b value images are more representative of true diffusion restriction and are relatively unaffected by factors like the perfusion. This has probably contributed to the higher specificity and lower sensitivity of the assessment system in our study. Most of the cancer sectors which were missed (received a PIRADSv2 assessment of 1 or 2) by mpMRI were low-grade cancers [79.5\% (n = 27/34)] and probably would not have been eligible for categorization as ‘clinically significant cancer’. Hence, PIRADSv2 alone proved to be an accurate method to diagnose clinically significant disease.