2398
Detecting resting-state brain functional networks using oxygen extraction fraction contrast1Center for MRI Research, Peking University, Beijing, China, 2MR Research China, GE Healthcare, Beijing, China
Synopsis
The traditional resting-state fMRI studies are based on the blood oxygenation level dependent (BOLD) contrast. Compared with BOLD, the oxygen extraction fraction (OEF) can more directly reflect the neuronal activities. However, due to the poor temporal resolution of existing OEF techniques, there is no study detecting resting-state networks with OEF contrast. In this study, the OEF contrast based resting-state networks were investigated through a newly proposed technique. Both seed-based correlation and independent component analysis were used and the results suggested that OEF can be used as an effective contrast to study resting-state brain networks.
Introduction
The blood oxygenation level dependent (BOLD) is often used as a contrast to investigate resting-state brain networks1-4. Because it is an indirect reflection of neuronal activity, the BOLD contrast shows limited spatial specificity. The oxygen extraction fraction (OEF), which detects the ratio of oxygen utilization and oxygen delivery, can potentially reveal higher spatial specificity of neuronal activities than BOLD. Detecting the spontaneous fluctuations of OEF signal in resting-state is crucial for understanding the underlying mechanism of brain functional networks. However, none functional connectivity studies have been investigated due to the relative poor temporal resolution of OEF mapping. In this study, a recently proposed technique5 was used to mapping OEF based resting-state networks with the temporal resolution of 3 seconds. Both seed-based functional connectivity (FC) and the group independent component analysis (gICA) were conducted to analyze OEF weighted time series to reveal resting-state brain networks.Methods
Twenty healthy subjects were scanned on a 3.0 T GE MR750 scanner equipped with an 8ch head coil. Every subject underwent a resting-state scan using the new sequence with eye closed to get the voxel-wise OEF time courses of the whole brain. The imaging parameters were as follows: repetition time (TR)=3000ms, imaging slices=22, filed of view (FOV)=26*26cm2, slice thickness=6mm, matrix size=64x64. The total scan time was 10 min and 30s. After the resting scan, 1 mm isotropic 3D T1-weighted images were acquired as the anatomic reference. (i) For seed-based FC analysis, the OEF value of each volume was calculated after head motion correction, spatial normalization and nuisance covariates regression of the six motion parameters. Then, nuisance covariates regression (including the mean time courses of white matter and cerebrospinal fluid), linear trend regression, band-pass filtering (0.01-0.08Hz) and spatial smoothing with a 6-mm Gaussian kernel were performed for all the OEF volumes. Then, the posterior cingulate cortex (PCC) was defined as a seed to conduct whole brain correlation analysis. At last, one-sample t-test was performed to detect cortical regions significantly correlated with PCC (FWE corrected p < 0.05). (ii) For gICA analysis, the OEF value of each volume was calculated after head motion correction, spatial normalization, spatial smoothing with a 6-mm Gaussian kernel and linear trend regression. The gICA analysis was performed using MELODIC tool implemented in the FMRIB Software Library (FSL). Then, the resting-state OEF data from twenty subjects were concatenated together and decomposed into 20 components.Results
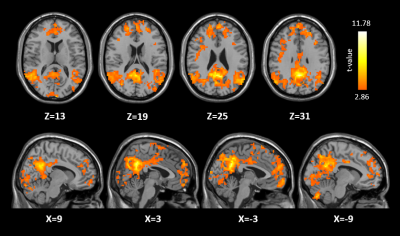
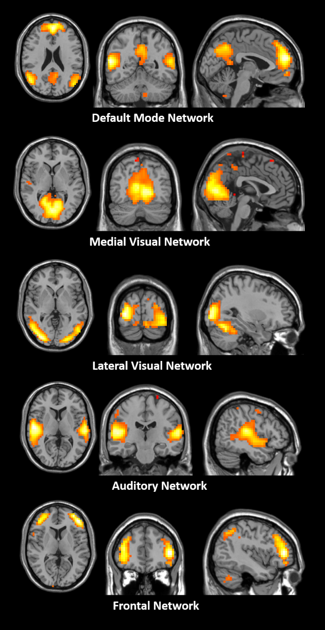
(i) Dynamic OEF fluctuations of the PCC were significantly correlated with brain regions in the default mode network (DMN) including medial prefrontal cortex (MPFC), bilateral angular gyrus and inferior parietal lobule as shown in Fig.1. (ii) As shown in Fig.2, there are five networks were identified as biologically meaningful based on the twenty components generated by gICA, including DMN, medial visual network, lateral visual network, auditory network and frontal network.Discussion and Conclusion
To our knowledge, this is the first investigation of OEF based resting-state brain networks. The DMN was revealed by both seed-based FC and gICA analysis. Some other networks were also revealed by gICA including medial visual network, lateral visual network, auditory network and frontal network. The seed-based FC networks are similar with those observed from BOLD, cerebral blood flow6, cerebral blood volume7 and cerebral metabolic rate of oxygen8. In conclusion, OEF can be used as an effective contrast to study brain networks in resting state, which is helpful to investigate the intrinsic physiological mechanism in the future.Acknowledgements
No acknowledgement found.References
1. Fransson P. Spontaneous low‐frequency BOLD signal fluctuations: An fMRI investigation of the resting‐state default mode of brain function hypothesis. Human brain mapping. 2005;26(1):15-29.
2. Fox MD, Snyder AZ, Vincent JL, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9673-9678.
3. Smith SM, Fox PT, Miller KL, et al. Correspondence of the brain's functional architecture during activation and rest. Proceedings of the National Academy of Sciences. 2009;106(31):13040-13045.
4. Smith SM, Nichols TE, Vidaurre D, et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nature neuroscience. 2015;18(11):1565-1567.
5. Yin Y, Zhang Y, Fan Y, et al. Simultaneous acquisition of oxygen extraction fraction and cerebral blood flow during brain activation. Proc Intl Soc Mag Reson Med. 2017;25:0359.
6. Zou Q, Wu CW, Stein EA, et al. Static and dynamic characteristics of cerebral blood flow during the resting state. Neuroimage. 2009;48(3):515-524.
7. Miao X, Gu H, Yan L, et al. Detecting resting-state brain activity by spontaneous cerebral blood volume fluctuations using whole brain vascular space occupancy imaging. NeuroImage. 2014;84:575-584.
8. Wu CW, Gu H, Lu H, et al. Mapping functional connectivity based on synchronized CMRO2 fluctuations during the resting state. Neuroimage. 2009;45(3):694-701.
Figures