2396
Using Temporal ICA to Selectively Remove Global Noise While Preserving Global Signal in Functional MRI Data1Washington University in St. Louis, Saint Louis, MO, United States, 2St. Luke's Hospital, Saint Louis, MO, United States, 3University of Oxford., Oxford, United Kingdom, 4Yale University, New Haven, CT, United States
Synopsis
A major unresolved methodological issue in fMRI is how to address the problem of spatially global noise, particularly in resting state functional connectivity data. Global signal regression is effective at removing global noise, which largely arises from physiological sources; however, it has the drawback of additionally removing global or semi-global neural signal as well. Here we present a method to selectively remove global noise while preserving global neural signal using temporal ICA. Thus, we remove a global positive bias in functional connectivity without inducing the network-specific negative bias that results from global signal regression.
Introduction
Temporal fluctuations in fMRI have successfully been used to study brain activity and connectivity for over two decades. Unfortunately, fMRI data also contain structured temporal “noise” from a variety of sources, including subject motion, subject physiology, and the MRI scanner. Recently, methods have been developed to automatically and selectively remove spatially specific structured noise from fMRI data using spatial Independent Components Analysis (ICA) and machine learning classifiers. Spatial ICA (sICA) is particularly effective at removing spatially specific structured noise from high temporal and spatial resolution fMRI data of the type acquired by the Human Connectome Project (HCP) and similar studies. However, sICA is unable (mathematically, by design) to separate spatially widespread “global” structured noise—e.g., blood flow modulations from subject respiration—from fMRI data. No methods currently exist to selectively and completely remove global structured noise while retaining global and semi-global signals related to neural activity. This has left the field in a quandary—to do or not to do global signal regression—given that both choices have substantial downsides1.
Methods
449 subjects from the young adult Human Connectome Project were scanned with resting state (rfMRI) and task-based fMRI (tfMRI) at 2mm isotropic with TR=0.72ms for 4800 frames of rfMRI and 3880 frames of tfMRI (7 tasks). The data were preprocessed according to the HCP-Style approach including correction of all imaging distortions, realignment for motion within and between modalities, and projection to a standard CIFTI grayordinates space using areal feature-based surface registration1,2,3,4. The data were first cleaned using a previously published sICA-based method (sICA+FIX) for spatially specific artifacts5.
Group sICA was then performed at dimensionality 137 and individual subject spatial maps and component timeseries generated using weighted regression2. The individual subject component timeseries were temporally concatenated and temporal ICA (tICA) was run on them. 84 reproducible rfMRI and 70 tfMRI tICA components were found, and were manually classified into signal vs noise components. The noise tICA component timecourses were regressed out of the sICA+FIX cleaned data.
We compared results of tfMRI and rfMRI analyses of sICA+FIX cleanup alone, sICA+FIX + tICA cleanup, and sICA+FIX + MGTR (Mean Gray Timecourse Regression, which is the grayordinate space analog to global signal regression). tfMRI analysis was performed using a grayordinates enabled FSL-based pipeline across the HCP’s 7 tasks6. rfMRI analyses included grayordinate-wise functional connectivity and connectivity gradient analyses and parcellated connectome analyses using full and partial correlation1,2.
Results
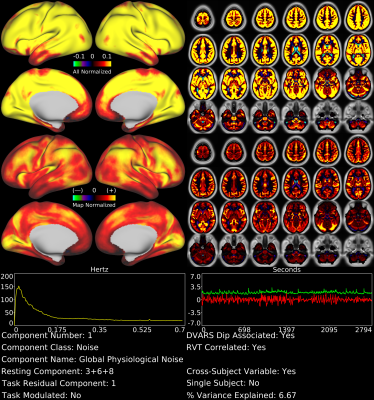
With tICA, we identified spatially global or semi-global components not discernible using sICA. For example, Figure 1 shows a global, gray matter specific tfMRI component that is the strongest component by temporal ICA variance explained (6.7%) and also shows the highest correlation of all components with the respiratory measure RVT7 (r=0.30). This component likely represents global fluctuations in BOLD signal induced by variations in respiration rate and depth. In the rfMRI data, respiration-related signal was split into 3 components, which together explained 8.5% of the temporal ICA variance and whose timeseries sum was also highly correlated with RVT (r=0.28).
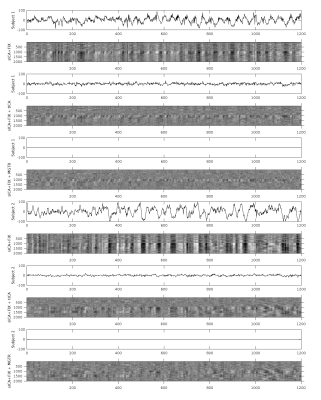
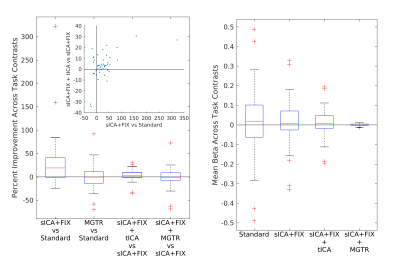
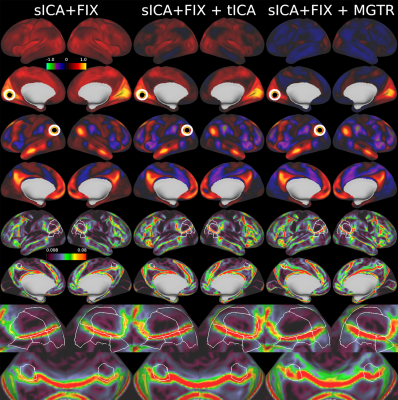
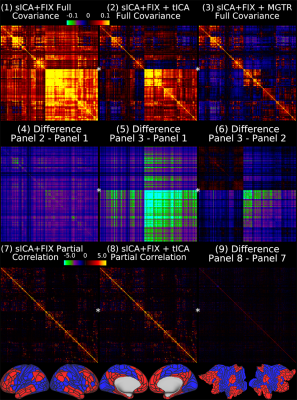
Overall, 25 tfMRI and 32 rfMRI components had properties consistent with structured noise, and 45 tfMRI and 52 rfMRI components had properties consistent with neural signal. Figure 2 shows that removing the structured noise tICA components is as effective as MGTR at removing global intensity variations from grayplots8 of rfMRI data, but without removing the whole global signal. Figure 3 shows that sICA+FIX and sICA+FIX + tICA are more beneficial to task analysis than are MGTR or sICA+FIX + MGTR, while avoiding the MGTR bias towards zero in the tfMRI contrast beta maps. Figure 4 shows the effects of tICA cleanup and MGTR on grayordinate-wise functional connectivity maps and their gradients, illustrating that tICA avoids biases in functional connectivity and gradient location that arise when using MGTR. Figure 5 uses parcellated functional connectivity matrices to characterize the biases of MGTR more explicitly, showing that tICA cleanup removes a global positive bias in functional connectivity, but that MGTR goes further and removes additional connectivity from non-cognitive regions (lower right quadrant of connectivity matrices).
Conclusions
Here we show that tICA can selectively segregate and remove global structured noise while retaining global neural signal in both tfMRI and rfMRI data. Comparing results before and after tICA cleanup to those from global signal regression shows that tICA cleanup removes biases caused by global physiological noise without inducing biases of global signal regression. We propose that tICA cleanup provides a “best of both worlds” solution to the global signal and noise dilemma.Acknowledgements
Supported in part by the Human Connectome Project, WU-Minn-Ox Consortium (1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; the McDonnell Center for Systems Neuroscience at Washington University; and NIH F30 MH097312 (M.F.G.), RO1 MH-60974 (D.C.V.E.). Funding to SS, JB, SH gratefully acknowledged via Wellcome Trust strategic award 098369/Z/12/Z. The authors would also like to thank Andreas Bartsch for helpful references related to negative CSF correlations in global and semi-global components.References
1Glasser, M.F., S.M. Smith, D.S. Marcus, J.L. Andersson, E.J. Auerbach, T.E. Behrens, T.S. Coalson, M.P. Harms, M. Jenkinson, S. Moeller, E.C. Robinson, S.N. Sotiropoulos, J. Xu, E. Yacoub, K. Ugurbil, and D.C. Van Essen. 2016b. The Human Connectome Project's neuroimaging approach. Nature neuroscience. 19:1175-1187.
2Glasser, M.F., T.S. Coalson, E.C. Robinson, C.D. Hacker, J. Harwell, E. Yacoub, K. Ugurbil, J. Andersson, C.F. Beckmann, M. Jenkinson, S.M. Smith, and D.C. Van Essen. 2016a. A multi-modal parcellation of human cerebral cortex. Nature. 536:171-178.
3Glasser, M.F., S.N. Sotiropoulos, J.A. Wilson, T.S. Coalson, B. Fischl, J.L. Andersson, J. Xu, S. Jbabdi, M. Webster, J.R. Polimeni, D.C. Van Essen, and M. Jenkinson. 2013. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 80:105-124.
4Robinson, E.C., S. Jbabdi, M.F. Glasser, J. Andersson, G.C. Burgess, M.P. Harms, S.M. Smith, D.C. Van Essen, and M. Jenkinson. 2014. MSM: a new flexible framework for Multimodal Surface Matching. NeuroImage. 100:414-426.
5Salimi-Khorshidi, G., G. Douaud, C.F. Beckmann, M.F. Glasser, L. Griffanti, and S.M. Smith. 2014. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. NeuroImage. 90:449-468.
6Barch, D.M., G.C. Burgess, M.P. Harms, S.E. Petersen, B.L. Schlaggar, M. Corbetta, M.F. Glasser, S. Curtiss, S. Dixit, C. Feldt, D. Nolan, E. Bryant, T. Hartley, O. Footer, J.M. Bjork, R. Poldrack, S. Smith, H. Johansen-Berg, A.Z. Snyder, and D.C. Van Essen. 2013. Function in the human connectome: task-fMRI and individual differences in behavior. NeuroImage. 80:169-189.
7Birn, R.M., J.B. Diamond, M.A. Smith, and P.A. Bandettini. 2006. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 31:1536-1548.
8Power, J.D., M. Plitt, T.O. Laumann, and A. Martin. 2017. Sources and implications of whole-brain fMRI signals in humans. NeuroImage. 146:609-625.
9Matthew F Glasser, Timothy S Coalson, Janine D Bijsterbosch, Samuel J Harrison, Michael P Harms, Alan Anticevic, David C Van Essen, Stephen M Smith. PrePrint. Using Temporal ICA to Selectively Remove Global Noise While Preserving Global Signal in Functional MRI Data. bioRxiv 193862; doi: https://doi.org/10.1101/193862
Figures