2393
High-Performance Correlation and Mapping Engine for Brain Connectivity Networks from High Resolution fMRI Data1Department of Electrical and Computer Engineering, Texas A&M University, College Station, TX, United States, 2Department of Psychological and Brain Sciences, Texas A&M Institute for Neuroscience, Texas A&M University, College Station, TX, United States
Synopsis
Seed-based Correlation Analysis (SCA) of fMRI data has been used to create brain connectivity networks. With close to a million unique voxels in a fMRI dataset, the number of calculations involved in SCA becomes high. With the emergence of the dynamic functional connectivity analysis, and the studies relying on real-time neurological feedback, the need for rapid processing methods becomes even more critical. This work aims to develop a new approach which produces high-resolution brain connectivity maps rapidly. Preliminary results show that this process can improve processing by a factor of 27 or more over that of a conventional PC workstation.
Introduction
Neuroimaging methods and analyses have rapidly evolved over the last decade, spurred by large collaborative funding initiatives such as the Human Connectome Project (HCP) in the US and the Human Brain Project in the EU. A major focus of these initiatives has been mapping and interpreting the connectivity of the brain. Understanding the wiring of the brain offers exciting possibilities to understand brain functions and aid in early detection and treatment of neurological diseases such as Alzheimer's, addiction, schizophrenia, dyslexia, autism, and ADHD [1].
Processing and analyzing these large datasets to determine the correlation between regions of the brain and functional tasks to generate a neurological connectivity map is computationally intensive. If each sampled voxel is used as a node (seed), generating a complete correlation map of a single human brain using straightforward correlation methods would require more than 120 hours of computation time on a multicore CPU computer [2, 3, 4].
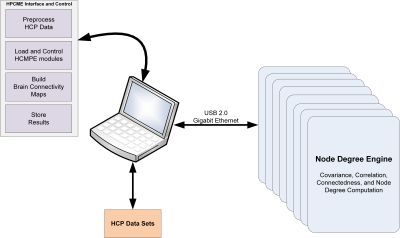
This research presents a High-Performance Correlation and Mapping Engine (HPCME) that enables high throughput processing of brain connectivity data sets. The HPCME system is an architecture that consists of a host computer, high-performance FPGA clusters and memory optimized for computing high volume correlation data.
Methods
The HPCME was designed to utilize a modular system comprised of many independent blocks calculating their assigned seed area of the same preprocessed, and organized voxel data sets. Each modular block had a parallel computation core and an independent and dedicated memory system to enable the near continuous streaming of the voxels and seeds being processed using correlation methods.
The overall architecture, shown in Figure 1, of the HPCME system, utilizes an FPGA system connected to a PC workstation. The PC workstation preprocesses, controls, and generates the final graphical brain connectivity map from the FPGA subsystem. The FPGA subsystem, known as the Node Degree Engine (NDE), was designed to process the voxels and seed data, and return data to generate a brain connectivity map. The NDEs can be scaled up so a cluster of FPGAs can run simultaneously.
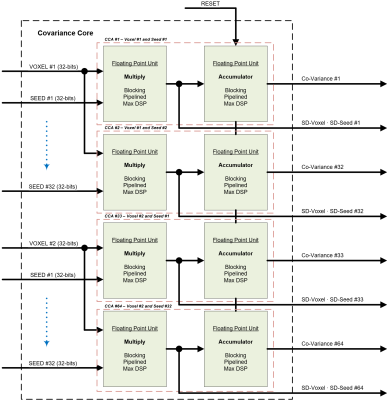
The Covariance Processing Core (CPC) is illustrated in Figure 2. The purpose of the core is to compute the covariance between the voxels and seeds. There are 64 processing elements in the core, and each has one floating point multiplier and one accumulator. The resulting correlation vectors is stored when a computed correlation value exceeds the given coefficient.
Results and Discussion
A representative set of preliminary results was generated with the Zynq 7Z030 SoC FPGA with the data being streamed at a conservative 200 MHz clock rate, with just one NDE core. A set of sixteen tests were conducted with similarly sized synthetic HCP data sets. These datasets contained 1,059,968 voxels, doing of the assigned seeds, and with 1,024 samples each leading to 5.617 x 1011 possible node connections. Also, the correlation coefficient was chosen to provide an 8% connectivity, versus the normally expected 2% to 3%.
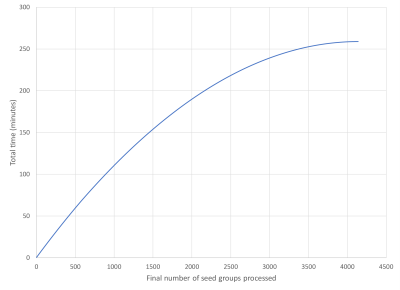
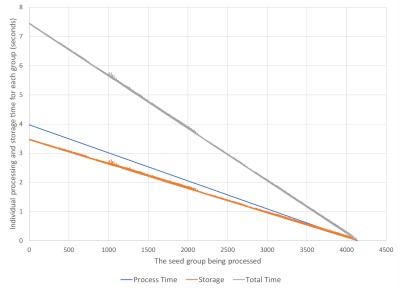
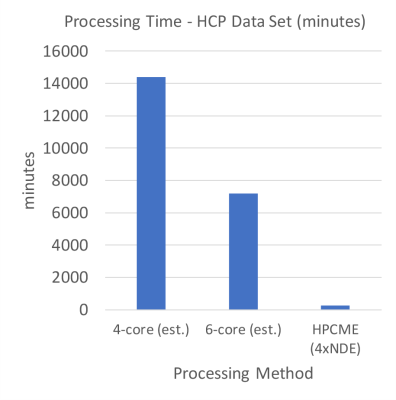
Figure 3 shows the total processing and storage time for the seed groups. The total time increases monotonically until the final group is processed. In Figure 4 the individual time of processing and the storage of the results vectors to the FLASH media is shown. This shows that as the processing group increments the process and storage time decreases. In Figure 5 we compare the HPCME results based upon using four NDEs, given the performance of our single NDE test, and estimated CPU performances for a full dataset [2, 3, 4]. Our estimates show that we will have a 27 times performance improvement over traditional CPU processing.
Conclusion
This research developed a new method to produce high-resolution brain connectivity maps rapidly. The new approach accelerates the correlation processing by using an architecture that includes clustered FPGAs and an efficient memory pipeline, which is termed High-Performance Correlation and Mapping Engine (HPCME). The method has been tested with datasets from the Human Connectome Project. The results showed that HPCME with four FPGAs can improve the SCA processing speed by a factor of 27 or more over that of a PC workstation with a multicore CPU and produce a high-resolution brain connectivity map in a matter of four hours, or less.
By reducing the processing time to generate brain connectivity maps, neuroscience researchers will be able to understand the various statistical links, thereby enabling them to be able to determine the effect of aging, Alzheimer’s, addiction, schizophrenia, dyslexia, autism, and ADHD. Also, being able to perform near real-time processing of fMRI data sets will enable DFC research to be conducted more rapidly over a larger segment number of nodes than currently [2].
Acknowledgements
Data was provided, in part, by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the US National Institute of Healths (NIH) Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
This work was partially supported, in part, by the US National Science Foundation (NSF) under the award number 1606136. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect those of the NSF.
References
[1] F. X. Castellanos, A. D. Martino, R. C. Craddock, A. D. Mehta, M. P. Milham, Clinical applications of the functional connectome, NeuroIm- age 80 (Supplement C) (2013) 527 – 540, mapping the Connectome.
[2] D. Akgn, nal Sakolu, J. Esquivel, B. Adinoff, M. Mete, Gpu accelerated dynamic functional connectivity analysis for functional mri data, Comput- erized Medical Imaging and Graphics 43 (Supplement C) (2015) 53 – 63.
[3] L. Minati, M. Cercignani, D. Chan, Rapid geodesic mapping of brain functional connectivity: Implementation of a dedicated co-processor in a field-programmable gate array (fpga) and application to resting state functional mri, Medical Engineering and Physics 35 (10) (2013) 1532 – 1539.
[4] L. Minati, D. Zac, L. DIncerti, J. Jovicich, Fast computation of voxel-level brain connectivity maps from resting-state functional mri using l1-norm as approximation of pearson’s temporal correla- tion: Proof-of-concept and example vector hardware implementa- tion, Medical Engineering and Physics 36 (9) (2014) 1212 – 1217.