2200
Magnetic susceptibility source separation using multi-echo GRE data only1Electrical and Computer Engineering, Seoul National University, Seoul, Republic of Korea, 2Department of Radiology, Seoul St. Mary's Hospital, Colleg of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
Synopsis
In this work, we explored an alternative approach of using nominal $$$R_2^{\:'}$$$ instead of measured $$$R_2^{\:'}$$$ in separating the two susceptibility sources. The linear relationship between $$$R_2^{*}$$$ and $$$R_2^{\:'}$$$ was investigated and used to obtain the nominal $$$R_2^{\:'}$$$ values. The positive and negative magnetic susceptibility source maps using nominal $$$R_2^{\:'}$$$ showed similar susceptibility distribution to the map using measured $$$R_2^{\:'}$$$.
Introduction
Recently, a new QSM algorithm that separates positive and negative susceptibility sources was proposed [1]. The method requires the estimation of $$$R_{2}^{\:'}$$$ which is defined as $$$R_{2}^{*}-R_{2}$$$ and, therefore, needs $$$R_{2}$$$ measurement. However, acquiring $$$R_{2}$$$ requires long scan time (10 min) and processing time (up to several hours) [2] and is not commonly performed in neuroimaging. In this work, we explored an alternative approach of using nominal $$$R_{2}^{\:'}$$$ instead of measured $$$R_{2}^{\:'}$$$ (= $$$R_{2,measured}^{\:'}$$$) in susceptibility source separation. The method was applied to healthy volunteers and multiple sclerosis (MS) patients and the resulting susceptibility maps were compared with those from $$$R_{2,measured}^{\:'}$$$.
Methods
[Estimating Nominal $$$R_{2}^{\:'}$$$]
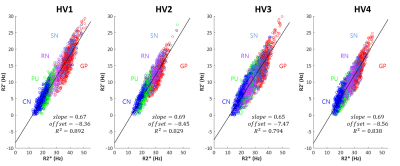
It has been shown that both $$$R_{2}^{\:'}$$$ and $$$R_{2}^{*}$$$ can be modeled as a linear equation to tissue susceptibility concentration [3-5]. Therefore, we can infer that the relationship between $$$R_{2}^{\:'}$$$ and $$$R_{2}^{*}$$$ is also linear. It can be written as follows:$$R_{2}^{\:'} = a\cdot{R_{2}}^{*}+b\:\:\:\:\:\:Eq. [1]$$ Once the two coefficients, $$$a$$$ and $$$b$$$, are known, we can use $$$R_{2}^{*}$$$ to estimate a nominal value of $$$R_{2}^{\:'}$$$. To obtain the coefficients, two different approaches are tested. First, the measurements in a literature [3] lead to $$$a=0.78$$$ and $$$b=-6.07$$$. The estimated $$$R_{2}^{\:'}$$$ using these values is referred to as $$$R_{2,nominal,lit.}^{\:'}$$$ hereafter. The second approach is to acquire $$$R_{2}^{*}$$$ and $$$R_{2,measured}^{\:'}$$$ from in-vivo data and then generate the coefficients using linear regression. This $$$R_{2}^{\:'}$$$ is referred to as $$$R_{2,nominal,fit}^{\:'}$$$.
[Experiments and data processing]
Four healthy volunteers and three MS patients were scanned at 3T. For $$$R_{2}^{*}$$$, gradient-echo was acquired (healthy volunteers: resolution=1×1×2mm3, TR=53ms, and TE=5.1:5.0:30.0ms; MS patients: resolution=0.5×0.5×2mm3, TR=53ms, and TE=5.8:6.2:36.7ms). To estimate $$$R_{2,measured}^{\:'}$$$, $$$R_{2}$$$ was acquired using multi-echo spin-echo (healthy volunteers: resolution=1×1×2mm3, TR=2400ms, and TE=10:10:100ms; MS patients: resolution=0.5×0.5×2mm3, TR=1800ms, and TE=10:10:90ms). An $$$R_{2}^{*}$$$ map was generated by mono-exponential fitting to the multi-echo data. An $$$R_{2}$$$ map was generated after $$$EPG_{SLR}$$$ correction [2]. $$$R_{2}^{\:'}$$$ was calculated by subtracting $$$R_{2}$$$ from $$$R_{2}^{*}$$$. The positive and negative susceptibility maps were acquired using [1].
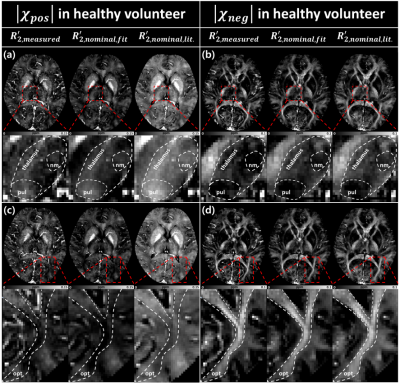
To estimate the coefficients of $$$R_{2,nominal,fit}^{\:'}$$$, a linear regression between $$$R_{2}^{*}$$$ and $$$R_{2,measured}^{\:'}$$$ was performed for voxels in five regions (SN, RN, CN, GP and PU) in healthy volunteers. No white matter region was included as $$$R_{2}^{*}$$$ has been shown to depend on not only susceptibility concentration but also fiber orientation [6, 7]. Outliers were removed using Cook’s distance [8]. The positive and negative susceptibility source maps from $$$R_{2,measured}^{\:'}$$$, $$$R_{2,nominal,fit}^{\:'}$$$, and $$$R_{2,nominal,lit.}^{\:'}$$$ were generated and compared. Thalamus, which includes iron-rich but myelin-lacking nuclei (pulvinar and nucleus medialis), and optic-radiation, which has myelin-rich but iron lacking region, were inspected [9-11]. The results were also compared in MS lesions.
Results
When relationship between $$$R_{2}^{*}$$$ and $$$R_{2}^{\:'}$$$ was explored, it showed a good linear relation (Fig. 1), agreeing with previous studies [3-5]. The slopes and offsets in [Eq. 1] of four healthy volunteers have similar values with small standard deviation ($$$slope=0.675\pm0.021$$$ and $$$offset=-8.21\pm0.499$$$). Hence, we used $$$R_{2,nominal,fit}^{\:'}=0.675\cdot{R_{2}}^{*}-8.21$$$ hereafter. From the literature [3], we found $$$R_{2,nominal,lit.}^{\:'}=0.78\cdot{R_{2}}^{*}-6.07$$$. The positive and negative susceptibility source maps of a healthy volunteer are shown in Fig. 2. Overall, the maps show similar contrast distribution. However, the positive susceptiblity map using $$$R_{2,nominal,lit.}^{\:'}$$$ was slightly overestimated (Fig. 2a and c right column). When thalamus was zoomed in, pulvinar and nucleus medialis are delineated in the negative susceptibility maps using $$$R_{2,measured}^{\:'}$$$ and $$$R_{2,nominal,fit}^{\:'}$$$ images (Fig. 2a and b left column and middle column).
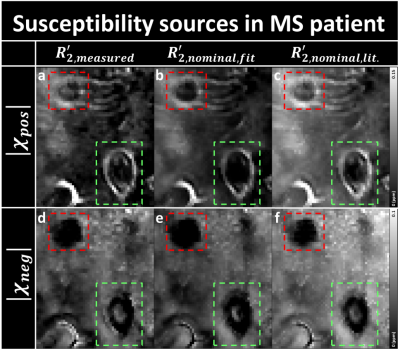
In MS patient results, all the positive maps show two ring-shaped lesions (Fig. 3), indicating iron accumulation at the rim of the lesions [12]. One of them shows potentially a feeding vein inside the lesion (Fig. 3; red box), which was still detectable but less apparent in the nominal $$$R_{2}^{\:'}$$$ corrected images. The negative map using $$$R_{2,measured}^{\:'}$$$ shows a demyelinated lesion (Fig. 3d and e; green box), and a potentially partially remyelinated lesion (Fig. 3d and e; red box). Similar appearances are observed in the negative susceptibility maps using $$$R_{2,nominal,fit}^{\:'}$$$, and $$$R_{2,nominal,lit.}^{\:'}$$$.
Discussion and Conclusion
In this work, we proposed a method to estimate nominal $$$R_{2}^{\:'}$$$ for the separation of magnetic susceptibility sources without measuring $$$R_{2}$$$. The resulting susceptibility maps using $$$R_{2,nominal,fit}^{\:'}$$$ showed similar distribution with those from $$$R_{2,measured}^{\:'}$$$ in healthy volunteers but the results are slightly different in MS lesions. On the other hand, when using $$$R_{2,nominal,lit.}^{\:'}$$$ for the separation, the susceptibility maps had higher values than those from $$$R_{2,measured}^{\:'}$$$ in both healthy volunteers and MS patients. The difference between $$$R_{2,nominal,fit}^{\:'}$$$ and $$$R_{2,nominal,lit.}^{\:'}$$$ may be explained by the different $$$R_2$$$ estimation process. To estimate $$$R_2$$$ reliably, we applied short echo spacing and $$$EPG_{SLR}$$$ correction which was not applied in [2].Acknowledgements
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI17C1919010017).References
[1] Lee, Jingu et al. “Separating positive and negative susceptibility sources in QSM.” ISMRM conference (2017).
[2] McPhee KC, Wilman AH. Transverse relaxation and flip angle mapping: Evaluation of simultaneous and independent methods using multiple spin echoes. Magn Reson Med 2017;77(5):2057-2065.
[3] Sedlacik, Jan, et al. "Reversible, irreversible and effective transverse relaxation rates in normal aging brain at 3T." Neuroimage 84 (2014): 1032-1041.
[4] C Langkammer, et al. “Quantitative MR imaging of brain iron: a postmortem validation study“, Radiology 257 (2010)
[5] Yao, Bing, et al. "Susceptibility contrast in high field MRI of human brain as a function of tissue iron content." Neuroimage 44.4 (2009): 1259-1266.
[6] Lee, Jongho, et al. "Sensitivity of MRI resonance frequency to the orientation of brain tissue microstructure." Proceedings of the National Academy of Sciences 107.11 (2010): 5130-5135.
[7] Lee, Jingu, et al. "An R2* model of white matter for fiber orientation and myelin concentration." NeuroImage 162 (2017): 269-275.
[8] Cook, R. Dennis. "Detection of influential observation in linear regression." Technometrics 19.1 (1977): 15-18.
[9] Duyn, Jeff H., and John Schenck. "Contributions to magnetic susceptibility of brain tissue." NMR in biomedicine 30.4 (2017).
[10] Bender, B., et al. Optimized 3D magnetization-prepared rapid acquisition of gradient echo: identification of thalamus substructures at 3T. American Journal of Neuroradiology 32.11 (2011): 2110-2115.
[11] Drayer, Burton, et al. Magnetic resonance imaging of brain iron. American journal of neuroradiology 7.3 (1986): 373-380.
[12] Zhang, Y., et al. "Quantitative susceptibility mapping and R2* measured changes during white matter lesion development in multiple sclerosis: myelin breakdown, myelin debris degradation and removal, and iron accumulation." American Journal of Neuroradiology 37.9 (2016): 1629-1635.
Figures