Danfeng Xie1, Li Bai1, and Ze Wang1,2
1Electrical and Computer Engineering, Temple university, Philadelphia, PA, United States, 2Department of Radiology, Temple university, Philadelphia, PA, United States
Synopsis
In
this study, we use Deep Learning-based (DL) method to denoising ASL CBF images.
Convolutional neural networks with a “wide” structure, residual learning and
batch normalization are utilized as the core of our denoising model. Comparing to
non-DL-based methods, the proposed method showed a significant SNR increase as well as partial volume effects improvement. Also, the DL-based method requires less
CBF input images, which significantly shorten the acquisition time and reduce
the chance of head motion.
Introduction
Cerebral Blood Flow (CBF) can be non-invasively measured with arterial
spin labeling (ASL) perfusion MRI but is subject to the low signal-to-noise-ratio(SNR)
[1]. Various methods have been proposed to denoise ASL MRI but only provide
moderate improvement. Deep learning (DL) is an emerging technique that can
learn the most representative signal from data without prior modeling and has
shown state-of-the-art performance on natural image denoising [2]. The purpose
of this study was to assess the feasibility and efficacy of DL in ASL MRI
denoising.Methods
ASL data were acquired from 280 subjects using a pseudo-continuous ASL sequence
(40 control/labeled image
pairs with labeling time = 1.48 sec, post-labeling delay = 1.5 sec, FOV=22 cm,
matrix=64x64, TR/TE=4000/11 ms, 20 slices with a thickness of 5 mm plus 1 mm
gap). CBF
images were calculated and spatially normalized into the MNI space using ASLtbx
[3,4] and SPM12. The deep ASL CBF denoising model was based on 5 layers of Convolutional
Neural Networks (CNNs) [5]. CNNs with a “wide” structure, meaning relatively
larger filter size (7x7) and more filters (128) was used for two reasons:
larger filters can better utilize spatial correlation among neighboring voxels
and more filters are able to capture the pixel-level distribution information more
effectively [2]. The network was implemented using the Wider Inference Network
(WIN) with no signal pooling and no fully connected output layer as often used
in regular CNNs. Grey matter (GM) probability map was incorporated as a
regularizer because CBF map shows a similar image contrast to that of a grey
matter map. To fasten and stabilize model learning, residual learning and batch
normalization were both included. The corresponding DL-based ASL denoising
model was dubbed as ASLDLD thereafter. To maximally show the benefit of ASLDLD,
we took the mean of first 10 CBF images without smoothing (meanCBF-10) as the input
image while used the mean of all 40 CBF images (meanCBF-40) with smoothing and
adaptive outlier cleaning [4] as the reference. The ASLDLD was trained with
data from 240 subjects’ 3D CBF maps (input and reference). The remained 40
subjects were used as test samples. Results
ASLDLD was compared with current non-DL-based denoising methods
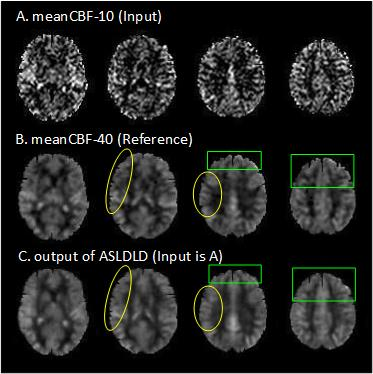
regarding the SNR of the resultant CBF images. Figure 1 shows the resultant CBF
maps from one representative subject. ASLDLD yielded superior performances to non-DL-based
methods. Especially, ASLDLD recovered CBF signal in the air-brain boundaries as
marked by the green boxes and signal loss due to partial volume effects as
labeled by the yellow boxes. Slightly better texture was obtained by ASLDLD
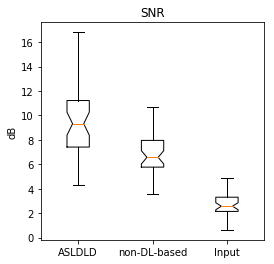
too. Figure 2 shows the SNR performance of different methods. SNR was
calculated as the ratio between the mean signal of a GM region-of-interest
(ROI) and the standard deviation of a white matter ROI. Compared to the non-DL-based
methods, ASLDLD showed a 38.6% SNR increase (p=1.14e-4). Discussion
In this study, we showed that DL-based denoising can substantially
improve ASL CBF SNR as well as the partial volume effects even only used the
mean CBF map of 10 pairs of ASL control/label image acquisitions. In other
words, ASLDLD can be utilized to significantly shorten the typical 5-6 mins acquisition
time by 75%, which would substantially reduce the chance of head motions, a big
confound in ASL imaging.Acknowledgements
No acknowledgement found.References
[1] Detre etc. Magnetic resonance in medicine, 231:37-45,
1992. [2] Liu etc. arXiv:1707.05414 [3] Wang etc. Magnetic resonance imaging, 262:261-269, 2008. [4] Wang etc.
Magnetic resonance imaging, 3010:1409-1415,
2012. [5] Krizhevsky etc. Advances in
neural information processing systems, 1097-1105, 2012.