2135
Multi-sequence and Habitat-based Radiomics Analysis to Predict MGMT Promoter Methylation Status in Grade II-IV Gliomas Using Magnetic Resonance Imaging1Chinese Academy of Sciences, Beijing, China, 2Shanxi Medical University, Taiyuan, China
Synopsis
In this study, we performed multi-habitat and multi-sequence MRI radiomics to make preoperative prediction on MGMT promoter methylation in grade II-IV gliomas. Quantitative imaging features were extracted on each habitat from CE-T1WI, T2FLAIR and ADC maps to reveal the genetic heterogeneity of the tumor and describe the subtle textural characteristics of different molecular subtypes. The habitat-integrated radiomics signature behaved more stable and had better predictive efficacy than one-region based radiomics signature. The final constructed predictive model incorporating the proposed radiomics signature and traditional clinical predictors achieved the optimal performance on the MGMT status.
Introduction
Gliomas are the most common central nervous system tumors with poor prognosis1-2. The average survival time ranged from 12 months to 5 years, making it a serious malignancy threatening people's health and life quality2. Fortunately, we figured out a subset of patients with the oxygen 6-methylguanine-DNA methyltransferase (MGMT) promoter methylation had better response to chemotherapy and performed longer survival3-5. studies revealed the clinical significance of MGMT methylation as a judicious marker for gliomas therapy decision making. However, MGMT status currently could only be obtained by large tissue sampling, which hinders to make timely therapy regime before the surgery6,7. In this study, we used radiomics as a method to solve this problem. Magnetic resonance images were utilized to reveal the relationship between the tumor heterogeneity and the MGMT status through textural analysis8. Particularly, we explored habitat based radiomics analysis for the prediction9. Imaging feature were extracted from both the tumor and peritumoral edema regions on T1-weighted, T2-FLAIR and Diffusion-weighted images, respectively. Precise and quantitative prediction on MGMT methylation was achieved and the optimal MR sequence that has the best predictive performance has been screened out.Methods
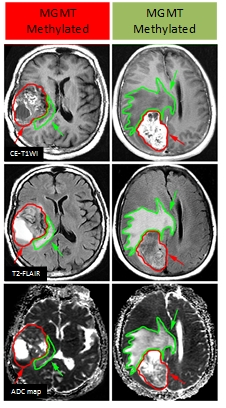
A cohort of 105 patients diagnosed with grade II-IV gliomas was enrolled in this study with institutional review board approval. CE-T1WI, T2FLAIR and DWI were performed with a 3.0-T MRI scanner (GE Signa HDxt, U.S.). We implemented manual segmentation on both tumor and peritumoral edema regions (Fig.1) of the three MR sequences to explore habitat-based radiomics. We calculated the intra-class correlation coefficient and concordance correlation coefficients to ensure the reproducibility and stability of the extracted features. Maximum relevance and minimum redundancy (mRMR) was used to select effective features with Bayesian information criteria as the stopping rule for the MGMT methylation predictive model construction. For each MR sequence, we established the radiomics signature using logistic regression modeling and the predictive performance of the signature was evaluated by receiver operating characteristics (ROC) curve analysis. Furthermore, we compared the predictive efficacy between the proposed radiomics signatures and traditional clinical factors using Delong test. The final predictive model incorporated both the useful clinical factors and the radiomics signature to achieve the best predictive accuracy.
Results
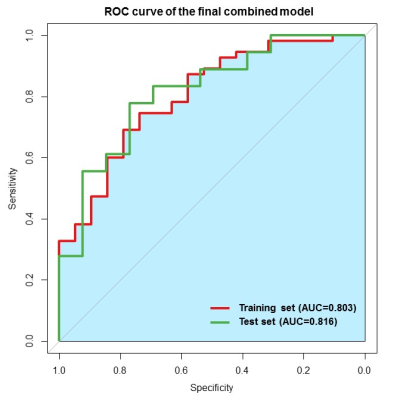
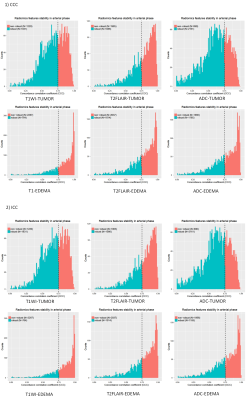
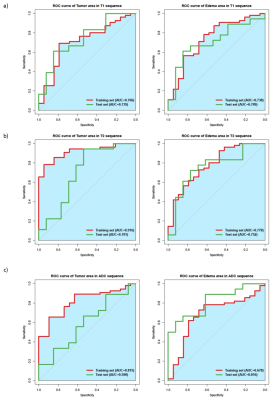
The final combined model achieved satisfied prediction on MGMT methylation status in grade II-IV gliomas with AUCs of 0.803 and 0.816 (Fig.2) on the training and validation datasets, respectively. For the habitat analysis, features extracted from tumor region had significantly superior stability and reproducibility than features extracted on peritumoral edema region (P < 0.05) (Fig.3). The multi-habitat radiomics signature performed better with AUCs of 0.793 and 0.812 on the training and validation datasets, respectively. It manifested the integration of the imaging features from different habitats improved the predictive efficacy on MGMT status compared with single radiomics signature (Fig.4). MR sequences T1WI and T2-FLAIR both had tantalizing ability to distinguish the methylation status on both tumor and peritumoral edema areas. However, DWI did not presented qualified prediction with AUC less than 0.7 on both the training and validation datasets. Clinical factor edema degree was selected using uni- and multi-variable analysis. The final combined model incorporated the multi-habitat and multi-sequence radiomics signature along with edema degree to fulfill the optimal predictive efficacy on MGMT methylation status (Fig.5).Discussion
This study reveals the intrinsic association between the imaging features and MGMT methylation status in grade II-IV gliomas. The prediction model was constructed based on the multi-habitat and multi-sequence radiomics features and effective clinical factors, providing reliable basis for the preoperative therapy decision making. It manifested improved performance for the habitat-based radiomics model than the traditional one-region based radiomics model. Habitats reflected detailed information on the micro-environment of the tumor lesion, which would assist describing the heterogeneity between different MGMT subtypes. The prior studies hold disparate opinions on the role of DWI radiological characteristics for MGMT prediction10-12. In this study, the DWI imaging features did not show valid predictive efficiency for the methylation distinguishment . T1WI and T2FLAIR sequences had superior performance and were used for the final predictive model construction. Edema degree was proved as the only resultful clinical factors for the prediction, which is in concordance with prior knowledge that MGMT methylated gliomas usually behaved with less peritumoral edema area.Conclusion
In this study, the preoperative prediction on MGMT methylation status in grade II-IV gliomas has been performed based on multi-habitat and multi-sequence radiomics. It enlightened the work to explore habitat analysis which is highly related to the micro-environment of the tumor and thus contain extra information useful for the molecular subtype prediction in gliomas.Acknowledgements
We are grateful to/for the following organizations/awards for funding this study: the National Natural Science Foundation of China (81227901 to Jie Tian); the International Innovation Team Program from the Chinese Academy of Sciences (20140491524 to Jie Tian); the Beijing Municipal Science and Technology Commission: Specific Projects Grant (Z151100001615012 to Jie Tian).References
1. States, C.B.T.R.o.t.U., Central Brain Tumor Registry of the United States Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004-2006. Hinsdale, IL, Central Brain Tumor Registry of the United States, 2010.
2. Wen, P.Y. and S. Kesari, Malignant gliomas in adults. New England Journal of Medicine, 2008. 359(5): p. 492-507.
3. Stupp, R., et al., Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine, 2005. 352(10): p. 987-996.
4. Cairncross, G., et al., Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. Journal of clinical oncology, 2012. 31(3): p. 337-343.
5. Hegi, M.E., et al., MGMT gene silencing and benefit from temozolomide in glioblastoma. New England Journal of Medicine, 2005. 352(10): p. 997-1003.
6. Sanai, N., J. Martino, and M.S. Berger, Morbidity profile following aggressive resection of parietal lobe gliomas. Journal of neurosurgery, 2012. 116(6): p. 1182-1186.
7. Tate, M.C., et al., Assessment of morbidity following resection of cingulate gyrus gliomas. Journal of neurosurgery, 2011. 114(3): p. 640-647.
8. Henson, J.W., P. Gaviani, and R.G. Gonzalez, MRI in treatment of adult gliomas. The lancet oncology, 2005. 6(3): p. 167-175.
9. Gillies, R.J., P.E. Kinahan, and H. Hricak, Radiomics: Images Are More than Pictures, They Are Data. Radiology, 2016. 278(2): p. 563.
10. Choi Y S, Ahn S S, Kim D W, et al. Incremental Prognostic Value of ADC Histogram Analysis over MGMT Promoter Methylation Status in Patients with Glioblastoma. Radiology, 2016, 281(1):175.
11. Romano A, Calabria L F, Tavanti F, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. European Radiology, 2013, 23(2):513-20.
12. Sunwoo L, Choi S H, Park C K, et al. Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semiquantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme.[J]. Journal of Magnetic Resonance Imaging, 2013, 37(2):351-8.
Figures