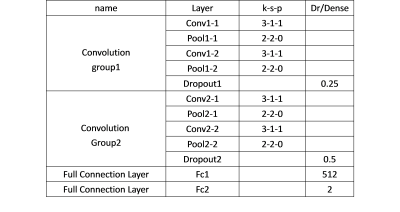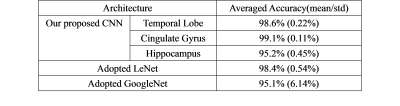1996
Detection of Alzheimer’s Disease Patients Based on Single Brain Region by Convolution Neural Networks1Harbin Institute of Technology (Shenzhen), Shezhen, China
Synopsis
Diagnosis plays an important role in preventing progress and treating the Alzheimer’s disease (AD). This paper proposed to predict the AD with a convolutional neural network (CNN), which can learn generic features capturing AD biomarkers. In particular, we extract some specific brain regions from structural MRI and apply MR features from the brain regions to detect AD patients in CNN framework, achieving accuracy up to 99% and outperforming some other classifiers from other studies.
Introduction
Deep learning technique has improved classification or detection performance in neuroimaging scenario during the past decade, indicating great potential application in clinical diagnostic trials. Researchers have achieved higher accuracy in detecting acute stroke patients based on MR diffusion images using convolution neural networks (CNN) compared to conventional statistical learning approaches 1. In contrast, it is still challenging to extend such techniques to Alzheimer’s disease (AD), as most neurodegenerative diseases lack obvious brain lesions on MR images. Several studies have established CNN-based pipeline to detect AD based on whole-brain structural, however, the association between image features derived from CNN and brain topological abnormality remains unclear. We hypothesize that MR features from specific local brain regions could also help detect AD patients with high accuracy in CNN framework. In current study, we applied the CNN approach on 3D T1 weighted images to detect AD patients based on a serious of single brain regions.Method
The structural MR images from Alzheimer’s disease Neuroimaging Initiative (ADNI) database were selected as input of our CNN pipeline since the brain local morphological features are our interests. Anatomical brain images were acquired on 1.5T MR scanners with MPRAGE sequence (TR=10ms, TE=4ms, matrix size=192×192, slice thickness=1.2). Two-class subjects with balanced sample size (110 healthy controls and 110 AD patients) were separately considered in this work. All the pre-processing steps of T1 weighted images for each subject were performed through a fully automated atlas-based parcellation platform (https://braingps.mricloud.org), including 1) bias correction, 2) normalization into standard space, 3) anatomical parcellation. For the output, the label maps covering the whole brain area were obtained for each subject by the method of atlas-bases segmentation 2, with each label corresponding to each brain parcel. All these above procedures have passed quality control tests by a neurologist. Based on our empirical knowledge, some brain parcels (including hippocampus, temporal lobe and cingulate gyrus) associated with AD pathology were selected as separate inputs of the following CNN pipeline. Each parcel consisting of multi-layer 2D images was obtained by parcel label mask, and rescaled into 128×128 dimension.
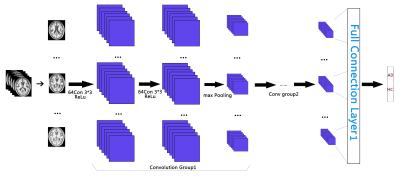
The proposed CNN architecture employed in current study includes two convolutional groups and full connection layers (see Table 1 and Figure 1). Each convolutional group is consisting of two convolutional layers with ReLU (Rectified Liner Units) 3 activation function in each layer, one pooling layer and one dropout layer. The CNN pipeline will finally generate the output percentage score as the classification probability belonging to AD or HC group for each input subject. Instead of random initialization of parameter adjustment, transfer learning 4 was introduced here to improve training efficiency. Namely, the pre-trained model of MICCAI challenge TADPOLE data 5 was employed as the parameter initialization of our CNN pipeline. In addition, Adam 6 was used in the following optimization procedure. In the data training processing, we randomly selected 25% of all subjects as training samples, and the other subjects as testing samples.
Result
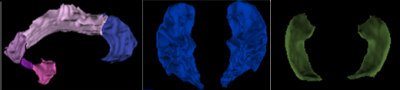
For better visualization effect, the surfaces of hippocampus, temporal lobe and cingulate gyrus from one healthy control subject are shown in 3D space (see Figure 2). The classification accuracy from validation test shows 98.6% for temporal lobe, 99.1% for cingulate gyrus and 95.2% for hippocampus, respectively. The accuracy of one previous whole-brain CNN model 7 is also listed as comparison (see Table 2).
Discussion
In this work, by establishing a CNN pipeline based on MR anatomical images, we successfully detected AD from healthy controls where an accuracy of 99.1% was achieved in the best-case scenario. Interestingly, the shift and scale invariant features extracted from multiple layers of CNN still work for single brain regions as the only input. These regional features are well congruent with previous findings that the brain structures (temporal lobe, cingulate cortex and hippocampus) largely affected by AD pathology can represent distinguished abnormality in terms of volume or morphology 8-10. By comprehensively combining image parcellation techniques and deep learning strategy together, the accuracy of our proposed AD detection model (best accuracy = 99.1%) precedes the previous model solely relying on deep learning (accuracy = 98.4%). Future work should focus on optimizing the selection criteria of local brain regions. In addition, our work also demonstrates high potential of using limited MR image slices to predict AD risk in future clinical diagnosis, as it allows to reduce MR scanning time.Acknowledgements
This work was made possible by grants from the Basic Research Foundation of Shenzhen Science and Technology Program (JCYJ20150403161923510), and the Basic Research Foundation Key Project Track of Shenzhen Science and Technology Program (JCYJ20160509162237418) and (JCYJ20170413110656460).References
1. Francis Dutil , Mohammad Havaei, Chris Pal , Hugo Larochelle, and Pierre-Marc Jodoin. A Convolutional Neural Network Approach to Brain Lesion Segmentation. Isles Challenge 2015, Germany.
2. J. P. Morin, C. Desrosiers and L. Duong, Atlas-based segmentation of brain magnetic resonance imaging using random walks. 2012 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Providence, 2012: 44-49.
3. Glorot X, Bordes A, Bengio Y. Deep Sparse Rectifier Neural Network. International Conference on Artificial Intelligence and Statistics. Cadiz, Spain. 2012.
4. S. J. Pan and Q. Yang, A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering. 2010; 10(22): 1345-1359
5. EuroPOND consortium funded by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 666992. 2017. TADPOLE: https://tadpole.grand-challenge.org
6. Diederik P. Kingma, Jimmy Ba, Adam: A Method for Stochastic Optimization, the 3rd International Conference for Learning Representations, San Diego, 2015
7. Saman Sarraf , Ghassem Tofighi. Classification of Alzheimer’s Disease Structural MRI Data by Deep Learning Convolutional Neural Networks. 22 Jul 2016. arXiv:1603.08631
8. M. Symms, et al. A review of structural magnetic resonance neuroimaging. Journal of Neurology. 2004; 9(75): 1235-1244.
9. R. Cuingnet, E. Gerardin, J. Tessieras, G. Auzias, S. Lehéricy, M.O. Habert, M. Chupin, H. Benali, O. Colliot. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage. 2011; 56: 766-781.
10. R. Alattas and B. D. Barkana. A comparative study of brain volume changes in Alzheimer's disease using MRI scans. 2015 Long Island Systems, Applications and Technology. Farmingdale, 2015:1-6.
Figures