1986
Automated Detection of Central Vessel Sign in Multiple Sclerosis using a 3D Deep Convolutional Neural Network1Radiology, Larner College of Medicine, University of Vermont, Burlington, VT, United States
Synopsis
A 3D deep convolutional neural network (dCNN) was trained to differentiate MS from non-MS lesions based on the orientation and location of a central vein ('central vein sign') relative to the lesion. Excellent performance was achieved using simulated FLAIR and T2*-weighted imaging, with realistic noise levels. The dCNN may be capable of identifying other discriminatory features from multimodal human imaging data.
Introduction
White matter lesions are the radiologic hallmark of multiple sclerosis (MS), but are not specific, and misdiagnosis continues to be problematic. Combining 3D FLAIR with T2*-weighted imaging ('T2*-FLAIR') allows for the identification of the presence of a central vein1. This central vessel sign (CVS) has high specificity for MS, but requires time-consuming manual evaluation of each lesion2. Our goal in this study is to determine if a 3D deep convolutional neural network can automatically learn to identify a central vein, and to evaluate its performance.
Methods
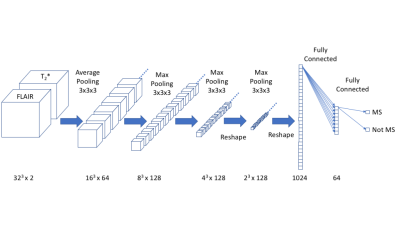
Deep convolutional neural networks (dCNNs) have been applied with great success to image recognition tasks3. We implemented a 3D dCNN with the architecture shown in Figure 1 using TensorFlow (Google). The network consists of alternating convolution and pooling layers, followed by two fully connected layers. The input to the network are two channels (volumes) of 32x32x32 voxels, corresponding to the FLAIR and T2*-weighted images.
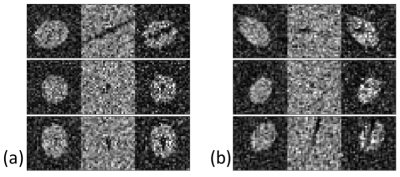
To demonstrate proof-of-concept, we trained the network based on 200 simulated 3D FLAIR and T2* images of artificial elliptical-shaped lesions, with added noise (SNR=2.5). Of these 200 pairs of images, 100 had a simulated vessel on the T2* image that ran close to the center of the lesion, angled within 5° of the central axis of the ellipsoid (MS group). In the remaining 100 pairs (non-MS), the vessel had a random orientation and location. Example images are shown in Figure 2. Neither the simulated FLAIR or T2* images individually provided discrimination between the two groups. To assess accuracy, 200 pairs of test and validation images were used.
Data augmentation was used to decrease the number of training lesions required. 1000 pairs of images were created by resampling from the original 200 with random spatial transformations. Independent testing and validation datasets (each consisting of 100 MS and 100 non-MS lesions) were used to evaluate the generalizable performance of the network.
Results
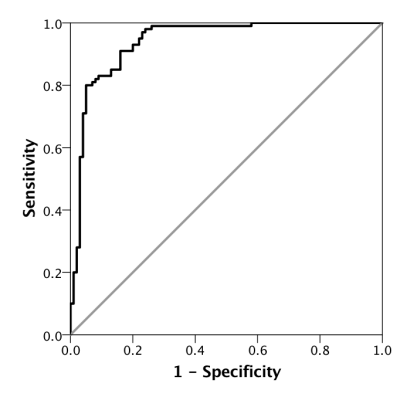
Over multiple training and testing runs, the accuracy on the testing and validation datasets were 0.896 ± 0.014 and 0.895 ± 0.040 respectively. An example of a receiver operator characteristic (ROC) curve is shown in Figure 3, with an area under the curve (AUC) of 0.940 ± 0.017. Training time was less than 10 minutes on an nVidia GTX 1080 Ti consumer GPU.
Discussion
This preliminary study used simulated data to investigate the feasibility of using a dCNN to differentiate MS lesions from non-MS lesions. In human subject data, lesions are unlikely to be perfectly elliptical, and often the central vein maybe difficult to identify.
However, the dCNN presented here has several advantages over a human reader:
- Use of the entire three-dimensional information that is available, rather than 2D sections.
- Natural extension to multimodal (multichannel) data. Combining FLAIR and T2* as a 'T2*-FLAIR' image is a convenient way for a human reader to interpret the spatial relationship between the vein and the lesion, but is unnecessary for the dCNN. Adding other modalities (such as T1-weighted images with or without contrast, QSM, etc) may improve performance.
- Fully automated, and extremely fast once trained.
- Capable of finding other features that distinguish MS from non-MS lesions. The current model was able to independently 'discover' that the spatial relationship between the lesion and vein could be used to separate MS and non-MS lesions. In human subject data, there may be information about, for example, the shape or texture of lesions that aids this discrimination.
In this study, only a modest number (200) of training lesions were used. Increasing the number of examples used for training will increase the generalizability and accuracy of the classification; with 1000 training lesions, a classification accuracy of over 95% was obtained. Our simulations were also conservative in that both the T2* and FLAIR images had SNR=2.5. In practice, both lesion and vessel conspicuity are likely to be higher, further increasing classification accuracy.
Conclusions
A 3D deep convolutional neural network was successfully trained to differentiate between simulated lesions with and without a central vein with high accuracy, even in data with poor SNR. Using multiple lesions from a single subject will result in higher diagnostic accuracy, even if the accuracy is only modest for a single lesion. A prior study using manual identification of central vein reported a sensitivity and specificity of of 0.81 and 0.83 respectively, based on analysis of three lesions from each subject.4 Future studies will investigate the diagnostic performance on human subject data using automatic lesion segmentation.5
Acknowledgements
Thanks to Safwan Wshah, Andrew Solomon, Josh Nickerson, and Risto Filippi for useful discussions on this work.References
1. Sati P, George IC, Shea CD, Gaitan MI, Reich DS. FLAIR*: a combined MR contrast technique for visualizing white matter lesions and parenchymal veins. Radiology. 2012;265(3):926-32.
2. Solomon AJ, Schindler MK, Howard DB, et al. "Central vessel sign" on 3T FLAIR* MRI for the differentiation of multiple sclerosis from migraine. Annals of clinical and translational neurology. 2016;3(2):82-7.
3. Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Commun Acm. 2017;60(6):84-90. doi: 10.1145/3065386. PubMed PMID: WOS:000402555400026.
4. Solomon AJ, Watts R, Ontaneda D, Absinta M, Sati P, Reich DS. Diagnostic Performance of Central Vein Sign for Multiple Sclerosis with a Simplified Three Lesion Algorithm. Submitted to Multiple Sclerosis Journal. 2017.
5. Shiee N, Bazin PL, Ozturk A, Reich DS, Calabresi PA, Pham DL. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 2010;49(2):1524-35.
Figures