1899
The diagnostic performance of DCE-MRI in glioma grading: A systematic review and meta-analysis1the first affiliated hospital of XI'AN jiaotong university, XI'AN, China
Synopsis
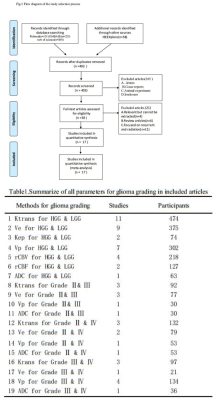
Different parameters of Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has been provided for noninvasive evluating gliomas pathology status. But the diagnostic performance of those parameters were variant among the recent reports during different type of gliomas. This study included 17 DCE-MRI studies regarding to differentiating different types of gliomas. The meta-analysis results demonstrated that Ve parameter of DCE-MRI has higher AUC in distinguishing HGGs from LGGs, gradeⅡ from grade Ⅲ and grade Ⅲ from gradeⅣ,respectively, Ktrans has higher AUC in distinguishing gradeⅡfrom grade Ⅳ; Among all the pamameters from DCE, Ktrans,Ve,Vp showed higher diagnostic performance in distinguishing different grade of gliomas.
Introduction
Accurate assessment of glioma grades is important which could determine the optimal therapy and prognosis[1].The method of dynamic contrast-enhanced magnetic resonance imaging(DCE-MRI) is widely researched in evaluating glioma grades recent years, but the diagnostic performance of those parameters were variant among the recent reports during different type of gliomas ,while this method is not extended to clinical practice[2]. Therefore, this study aims to systematically review and meta-analyze DCE-MRI for differentiating different grade of gliomas.Methods
This meta-analysis was performed in accordance with the PRISMA guidelines[3]. The Pubmed ,Web of Science, EMBASE and Google Scholar databases were systematically searched until 31 October 2017, using following terms combinations: DCE & glioma, dynamic contrast enhanced & glioma, Ktrans & glioma, Ve & glioma. Quality Assessment Tool for Diagnostic Accuracy Studies version 2(QUADAS-2)[4]was used by two independent authors to evaluated the quality of each included studies. The primary outcomes were Ktrans , Ve, Vp, Kep, rCBV, rCBF . The extent of heterogeneity was assessed using chi-square value test and inconsistency index (I2) of diagnostic odds ratio (DOR). A Spearman correlation coefficient was computed between logit of SEN and logit of (1−SPE) to assess threshold effect, P<0.05 indicated significant difference. The summary receiver-operating characteristic curve (SROC), area under the curve (AUC) and Q* index were calculated. The above mentioned statistical analyses were performed using Meta-DiSc statistical software version1.4 and Review manager 5.3[5].Publication bias was assessed by Deeks’ funnel plot. Formal testing for publication bias was conducted using a regression of the diagnostic log odds ratio against ESS1/2 (effective sample size) and weighting according to the effective sample size, with P<0.10 indicating significant asymmetry[6]. This statistical analysis was performed using Stata 12.0 software (StataCorp LP, College Station, TX, USA).Results
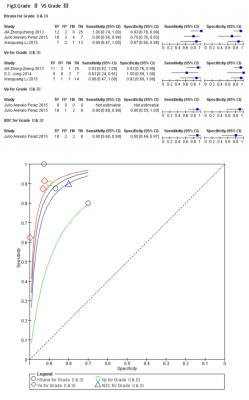
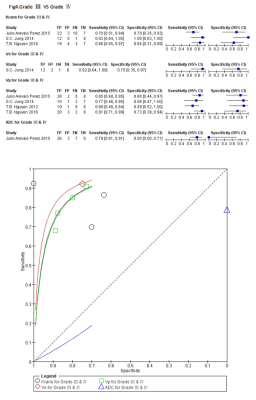
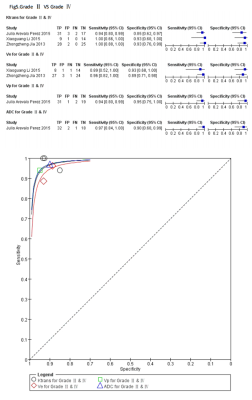
A flow diagram of the study selection is shown in Fig 1. 17studies[2,7-22]were included in our study. QUADAS-2 was used to assess the quality of each studies.474 patients were included and 19 comparison groups were performed in distinguishing different grade of gliomas (Table1).The various indicators obtained directly or indirectly from each study were calculated by a doctor who has four years experience. Ve has higher AUC in distinguishing HGGs from LGGs(Fig2), gradeⅡ from grade Ⅲ(Fig3) and grade Ⅲ from gradeⅣ(Fig4),respectively. Ktrans has higher AUC in distinguishing gradeⅡfrom grade Ⅳ(Fig5); Ktrans, Ve and Vp all have higher diagnostic performance in distinguishing HGGs from LGGs(Fig2).Discussion
DCE-MRI provides more truthful tumor perfusion information, Ktrans ,Ve and Vp are the most important quantitative parameters, which showed high diagnostic performance in this meta-analysis.
In this study ,Ve showed higher AUC in distinguishing HGGs from LGGs, gradeⅡ from grade Ⅲ and grade Ⅲ from gradeⅣ,respectively, which indicated that Ve is the most important parameter in glioma grading , this result consist with one previous study,which showed Ve is the only determining factor for glioma grading by the logistic regression[8]. Ktrans has a little lower AUC than Ve in distinguishing HGGs from LGGs and has higher AUC in distinguishing grade Ⅱfrom grade Ⅳ,which indicated that Ktrans is another important factor in glioma grading, because Ktrans could represent the tumorous microvascular permeability. However, one study showed that Ktrans,Ve,Kep and rCBF have no significant difference in distinguishing HGGs from LGGs, only Vp and rCBV showed media diagnostic performance[7], The main reason for this issue may explain as the following aspects. There is no standardization as to the optimal method of assessing perfusion[23], T1 time, cut-off value and pharmacokinetic model are different during included studies[2,7-22].These may exist heterogeneity and the results may untruthfulness. Nevertheless , the basic theory of methods were similar, the resultant error is not so large during closely acquisition times[2]. When using different DCE protocol for grading gliomas, parametric errors occur in the same direction, their resulting distributions could be unchanged and diagnostic utility is preserved. Therefore, our results are credible and valuable, meanwhile ,further investigation involving radiological-pathological correlation is needed to determine a standard in using DCE-MRI, in order to spread DCE-MRI to clinical practice.
There was also one studies compared DCE with other mehods (ADCmin) for distinguishing gliomas in our meta-analysis[13], although ADCmin showed higher sensitivity/specificity in distinguishing HGGs from LGGs, grade Ⅱ from grade Ⅲ, grade Ⅱ from gradeⅣand media sensitivity in grade Ⅲ from gradeⅣ, but the AUC of ADCmin was lower than Ktrans and Ve parameters.
Summarize
This meta-analysis provides evidence that different parameters of DCE-MRI could present different tumorous pathology status and have higher diagnostic performance in distinguishing different grade gliomas, which would contribute greatly to various clinical situations.Acknowledgements
This study was supported by the National Key Research and Development Program of China (2016YFC0100300), National Natural Science Foundation of China (No. 81471631, 81771810 and 51706178), the 2011 New Century Excellent Talent Support Plan of the Ministry of Education, China (NCET-11-0438) and the Clinical Research Award of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF-CRF-2015-004).References
[1] DeAngelis L M. Brain tumors[J]. New England Journal of Medicine, 2001, 344(2): 114-123.
[2] Abe T, Mizobuchi Y, Nakajima K, et al. Diagnosis of brain tumors using dynamic contrast-enhanced perfusion imaging with a short acquisition time[J]. SpringerPlus, 2015, 4(1): 88.
[3] Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med. 2009;151:W65–W94.
[4] Leeflang M M G, Deeks J J, Gatsonis C, et al. Systematic reviews of diagnostic test accuracy[J]. Annals of internal medicine, 2008, 149(12): 889-897.
[5] Zamora, J., Abraira, V., Muriel, A., Khan, K., &Coomarasamy, A. (2006). Meta-DiSca software for meta-analysis of test accuracy data. BMC medical research methodology, 6(1), 31.
[6] Deeks J J, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed[J]. Journal of clinical epidemiology, 2005, 58(9): 882-893.
[7] Gupta P K, Saini J, Sahoo P, et al. Role of Dynamic Contrast-Enhanced Perfusion Magnetic Resonance Imaging in Grading of Pediatric Brain Tumors on 3T[J]. Pediatric Neurosurgery, 2017, 52(5): 298-305.
[8] Brendle C, Hempel J M, Schittenhelm J, et al. Glioma Grading and Determination of IDH Mutation Status and ATRX loss by DCE and ASL Perfusion[J]. Clinical Neuroradiology, 2017: 1-8.
[9] Sahoo P, Gupta P K, Awasthi A, et al. Comparison of actual with default hematocrit value in dynamic contrast enhanced MR perfusion quantification in grading of human glioma[J]. Magnetic resonance imaging, 2016, 34(8): 1071-1077.
[10] Santarosa C, Castellano A, Conte G M, et al. Dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging for glioma grading: preliminary comparison of vessel compartment and permeability parameters using hotspot and histogram analysis[J]. European journal of radiology, 2016, 85(6): 1147-1156.
[11] Arevalo‐Perez J, Kebede A A, Peck K K, et al. Dynamic contrast‐enhanced MRI in low‐grade versus anaplastic oligodendrogliomas[J]. Journal of neuroimaging, 2016, 26(3): 366-371.
[12] Li X, Zhu Y, Kang H, et al. Glioma grading by microvascular permeability parameters derived from dynamic contrast-enhanced MRI and intratumoral susceptibility signal on susceptibility weighted imaging[J]. Cancer Imaging, 2015, 15(1): 4.
[13] Arevalo‐Perez J, Peck K K, Young R J, et al. Dynamic Contrast‐Enhanced Perfusion MRI and Diffusion‐Weighted Imaging in Grading of Gliomas[J]. Journal of Neuroimaging, 2015, 25(5): 792-798.
[14] Nguyen T B, Cron G O, Mercier J F, et al. Diagnostic accuracy of dynamic contrast-enhanced MR imaging using a phase-derived vascular input function in the preoperative grading of gliomas[J]. American Journal of Neuroradiology, 2012, 33(8): 1539-1545.
[15] Jia Z, Geng D, Xie T, et al. Quantitative analysis of neovascular permeability in glioma by dynamic contrast-enhanced MR imaging[J]. Journal of Clinical Neuroscience, 2012, 19(6): 820-823.
[16] Awasthi R, Rathore R K S, Soni P, et al. Discriminant analysis to classify glioma grading using dynamic contrast-enhanced MRI and immunohistochemical markers[J]. Neuroradiology, 2012, 54(3): 205-213.
[17] Zhao J, Yang Z, Luo B, et al. Quantitative evaluation of diffusion and dynamic contrast-enhanced MR in tumor parenchyma and peritumoral area for distinction of brain tumors[J]. PloS one, 2015, 10(9): e0138573.
[18] Jia Z, Geng D, Liu Y, et al. Low-grade and anaplastic oligodendrogliomas: differences in tumour microvascular permeability evaluated with dynamic contrast-enhanced magnetic resonance imaging[J]. Journal of Clinical Neuroscience, 2013, 20(8): 1110-1113.
[19] Jia Z Z, Geng D Y, Liu Y, et al. Microvascular permeability of brain astrocytoma with contrast-enhanced magnetic resonance imaging: correlation analysis with histopathologic grade[J]. Chinese medical journal, 2013, 126(10): 1953-1956.
[20] Nguyen T B, Cron G O, Bezzina K, et al. Correlation of Tumor Immunohistochemistry with Dynamic Contrast-Enhanced and DSC-MRI Parameters in Patients with Gliomas[J]. American Journal of Neuroradiology, 2016, 37(12): 2217-2223.
[21] Jung S C, Yeom J A, Kim J H, et al. Glioma: application of histogram analysis of pharmacokinetic parameters from T1-weighted dynamic contrast-enhanced MR imaging to tumor grading[J]. American Journal of Neuroradiology, 2014, 35(6): 1103-1110.
[22] Haris M, Gupta R K, Singh A, et al. Differentiation of infective from neoplastic brain lesions by dynamic contrast-enhanced MRI[J]. Neuroradiology, 2008, 50(6): 531.
[23] Cher Heng Tan1,Brian Paul Hobbs,Wei Wei, et al. Dynamic Contrast-Enhanced MRI for the Detection of Prostate Cancer: Meta-Analysis[J].AJNR,2015,204:w439-w448.
Figures