1806
Convolutional Neural Networks on Functional Connectivity Derived From r-fMRI: Explore the Effects of ThresholdsXingjuan Li1, Yu Li1, and Xue Li1
1School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
Synopsis
In this study, we propose a novel CNN to predict autism from functional brain networks. Experimental results demonstrate that the predictive ability of CNN outperforms a logistic regression method by 8% and a five-layer fully-connected network (FCN) by approximately 7%. Network thresholding is often used to control false connections arising in the process of constructing functional brain networks. We also compare the influence of different thresholds on the performance of proposed CNN. Experimental results show that CNN is robust to false connections. Our study will contribute to predict reliable clinical outcomes in autism using deep learning on brain networks.
Introduction
Brain connectivity is of great interest in the neuroscience community to study brain-behaviour relations. It refers to anatomical link of statistical dependencies or causal effects among distributed units in the brain [1]. The connections or interactions among brain regions are often modelled as complex networks, which are also known as brain networks. Topological studies of brain network reveal that normal human brain is associated with cost-efficiency, modularity and a rich club of connected hubs [2]. Disruptions of brain network have been found to be related with some psychological disorders, such as autism spectrum disorder (ASD). Brain networks are often been used as the data feature to classify diagnostic class or predict clinical factors, especially in the context of studying patients with cognitive deficits. Recently, deep learning opens up new frontiers to analyse medical data, leading to improved accuracy in assessing disease [4]. Success of deep learning may attribute to the multiple layers features learned by deep neural networks. However, deep learning is rarely applied to analyse brain networks because of their high-dimensional and irregular structured characteristics. In this study, we proposed a multiple layer convolutional neural network (CNN) to learn the network features for predicting diagnostic labels in individuals with ASD. Due to the choice of network construction methods, false connections often arise and influence the network analysis results [3]. Network thresholding is the most common method to control false connections. However, there is no standard method to choose an appropriate threshold. The threshold is often determined arbitrarily. In this paper, we investigate the influence of different thresholds on resting-state functional brain networks in classifying ASD from normal controls (NC) using the proposed CNN.Method
We constructed functional brain networks using resting-state functional magnetic resonance images (r-fMRI) collected from ABIDE database (http://fcon_1000.projects.nitrc.org/indi/abide/), including 177 ASDs, 209 NCs. Network construction steps include: (1) All images were pre-processed for denoising, extracting brain and normalization by FSL software (https://fsl.fmrib.ox.ac.uk/); (2) We extracted 116 brain regions using AAL template, and selected 108 regions from left and right hemispheres to construct the interhemispheric connectivity; (3) We estimated Pearson correlation of mean fMRI signals among all distributed brain regions. (4) Constructed functional brain networks were thresholded at 0.1, 0.2 and 0.3. We designed a five layer convolutional neural network, including two convolutional layers, two subsampling layers and one fully-connected (FC) layer. In order to learn the geometrical information of brain networks, we performed spectral graph convolution $$G×f=UT(UG⊗Uf)$$ at each convolutional layer. G is the graph representation of brain network, f is convolution filters, and U is unitary matrix. The CNN was trained using statistic gradient descent (SGD) method with a momentum of 0.9. In the training process, we deployed a 10-fold cross validation strategy to split the data into training data and test data. Our model was evaluated using accuracy, precision, and recall. We also compared our proposed CNN on original functional brain network with other machine learning methods, including logistic regression and a five FC neural network.Results
We first compare our model with logistic regression and a five-FC network (see table 1). As we can see from the table, our method significantly outperforms the other two methods in terms of the binary classification task. To be more specifically, the accuracy of proposed CNN is about 8% higher than the logistic regression, and 7% higher than a five-FC neural network. The results of different thresholds on the classification between ASDs and NCs have been listed in table 2. As shown in the table, the proposed CNN performs best in original functional networks and it is less likely to be affected by different thresholds.Discussion and Conclusion
Broadly, the proposed CNN performs well in distinguishing individuals with ASD from NCs. In addition, experimental results show that the proposed CNN is less likely to be affected by false connections in functional brain networks. Compared to the classification results reported using support vector machine (average accuracy of 73.89%) in [5], the CNN outperforms this approach by about 2%. We regard this work as a proof of idea for the following reasons. Firstly, the training data is relative small due to the limitation of available data. Additionally, the double-blinded experimental results are hard for both doctors and patients to accept. Even so, deep learning emerges as new frontiers to analysis brain networks with high accuracy.Acknowledgements
No acknowledgement found.References
[1] Bassett, D. S., & Sporns, O.. Network neuroscience. Nature neuroscience, 2017; 20(3), 353. [2] Bullmore, E. and Sporns, O.. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience, 2009; 10(3), pp.186-198. [3] Drakesmith, M., Caeyenberghs, K., Dutt, A., Lewis, G., David, A.S. and Jones, D.K.. Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. NeuroImage, 2015; 118, pp.313-333. [4] Greenspan, H., van Ginneken, B. and Summers, R.M.. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Transactions on Medical Imaging, 2016; 35(5), pp.1153-1159. [5] Plitt, M., Barnes, K. A., & Martin, A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical, 2015; 7, 359-366.Figures
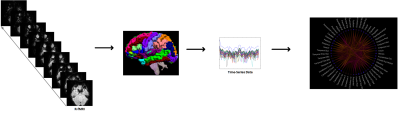
Fig. 1. Construction of functional brain networks. All images are parcellated using AAL template. Then, we extract 116 regions as network nodes. Edges are estimated using Pearson correlation between all pair-wise mean fMRI signals among distributed regions.
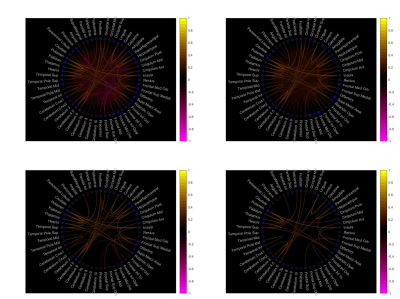
Fig. 2. Thresholding functional brain networks. On the top left is the original functional connections without thresholding, On the top right is the functional connections thresholded by 0.1. The bottom left and right functional connections are thresholded by 0.2 and 0.3 respectively.
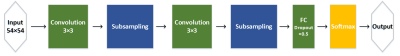
Fig. 3. The architecture of proposed CNN. It includes two convolutional layers, two subsampling layers, and one FC layers. We use dropout strategy in FC layer to control overfitting problem.
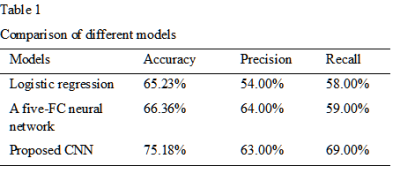
Table 1. Comparison of different models
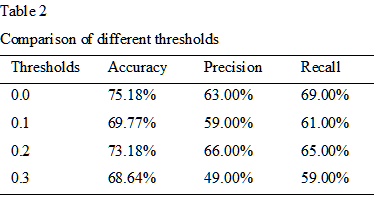
Table 2. Comparison of different thresholds on the performance of proposed CNN