1783
Comparison of Thalamus Segmentation Using Publicly Available Segmentation Methods in a Pediatric PopulationSalem Hannoun1, Rayyan Tutunji2, Maria El Homsi2, and Roula Hourany2
1Nehme and Therese Tohme Multiple Sclerosis Center, American University of Beirut Medical Center, Beirut, Lebanon, 2Radiology Department, American University of Beirut Medical Center, Beirut, Lebanon
Synopsis
107 subjects were recruited between the ages of one month and 18 years. The study aimed to investigate the differences in the accuracy of five publicly available segmentation techniques on T1-enhanced and non-enhanced images compared to manual segmentation of the thalamus in a pediatric population. volBrain had the best outcomes in enhanced and non-enhanced images. Image segmentation using volBrain is the ideal methodology for thalamus segmentation. Gadolinium-enhancement negatively affects the outcomes of all the tested automated segmentation.
Purpose
The manual tracing of subcortical gray matter such as the thalamus, requires a high level of expertise. Their involvement is increasingly recognized as an important pathophysiological feature. Several methods have been previously developed and introduced to perform automatic and semi-automatic regional segmentation as accurately and specifically as possible. Such tools accelerate data analysis in large studies, and deliver reproducible and consistent outcomes, which are crucial for obtaining reliable results (1). However, in several clinical studies, the lack of time and the cost of the MRI imply the acquisition of T1 after gadolinium (Gd) injections only. Gd signal makes it hard for automatic tools to segment brain regions. What could also affect the automatic tissue-classification in enhanced images is the variability over patients of the administered Gd dose, as well as the timing of contrast administration (2). We aimed to investigate the differences in the accuracy of publicly available segmentation techniques on T1-enhanced and non-enhanced images compared to manual segmentation of the thalamus in a pediatric population.
Methods
107 subjects were recruited between the ages of one month and 18 years. 3D T1 images were acquired on either a 1.5T or a 3T scanners (Ingenia, Phillips). Images were controlled for major artifacts that could implicate an error during segmentation, then classified in two groups: 3DT1 without (n=74) and with Gd administration (n=33). Manual segmentation of the thalamus was done by one rater, with another rater doing 33 measurements for inter-rater comparison. Automated segmentation on the same subjects was performed with volBrain, MRICloud, FSL Anat, FIRST, and FreeSurfer (Figure 1). Default parameters were used for all segmentation algorithms. A mask of the intersections between the manual and automated segmentation was created for each algorithm to measure the degree of similitude (DICE) with the manual segmentation. Interrater reliability for the manual segmentation performed by both raters was measured using a weighted Kappa. The similitude indexes were examined for general differences between the automated techniques using ANOVA. Differences between enhanced and non-enhanced T1 were studied via a t-test.
Results
We found that volBrain segmentation had the best outcome in terms of accuracy with regards to the manual segmentation with a DICE of 0.867 for non-enhanced T1 and 0.802 for Gd-enhanced T1 images (Figure 2). On the other end of the spectrum, MRICloud proved to have the lowest DICE in both enhanced (DICE=0.729) and non-enhanced images (DICE=0.712). FSL-Anat and FIRST came in second and third respectively. DICE scores were significantly higher in non-enhanced compared to enhanced images. Age was not a significant predictor of DICE in any of the measurements.
Conclusion
The implementation of automated techniques makes large scale populations studies much easier to conduct. Manual delineation of specific regions of interest or even whole brain segmentation could be tedious and time consuming. To this end, several segmentation techniques have been developed, each based on different algorithms, some being semi-automatic, others fully automatic. Among five automated segmentation techniques, volBrain proved to have the best outcomes in enhanced and non-enhanced MRI images. While most studies usually use T1-WI for structural analysis, it is often the case in retrospective studies that non-enhanced images are not available, as is the case with 30% of the subjects examined in our study. Therefore, there is a need to assess the extent of the effects of Gd enhancement on automated segmentation tools. Indeed, Gd-enhancement negatively affects the outcomes of all the tested automated segmentation. T1 non-enhanced image segmentation using volBrain would appear to be the ideal methodology for segmentation of the thalamus. This will achieve results closest to manual segmentation while reducing the amount of time and computing power needed by researchers.Acknowledgements
No acknowledgement found.References
- Mulder, E.R., de Jong, R.A., Knol, D.L., van Schijndel, R.A., Cover, K.S., Visser, P.J., Barkhof, F., Vrenken, H., Alzheimer’s Disease Neuroimaging Initiative, 2014. Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and FIRST. Neuroimage 92, 169–81.
- Warntjes, J.B.M., Tisell, A., Landtblom, A.-M., Lundberg, P., 2014. Effects of gadolinium contrast agent administration on automatic brain tissue classification of patients with multiple sclerosis. AJNR. Am. J. Neuroradiol. 35, 1330–6.
Figures
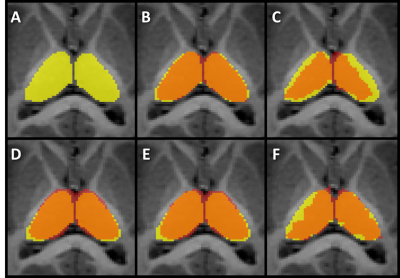
Image of the manually segmented Thalamus in yellow (A) superimposed on the
automatic segmentation done with volBrain (B), MRICloud (C), FSL Anat (D),
FIRST (E), and FreeSurfer (F). Areas in red represent voxels where no
intersection was seen between manual and automatic segmentations. Areas in
orange represent voxels where both manual and automatic segmentations
intersected.
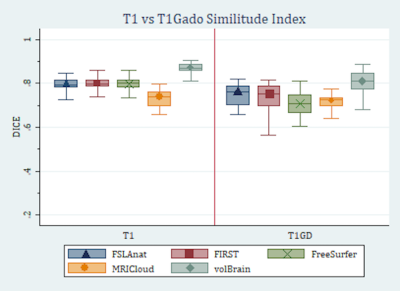
Box plot showing the
DICE for enhanced and non-enhanced images. The DICEs for each method are higher
in the non-enhanced images, with the whiskers showing less variation as well.
This indicates more accuracy and consistency in non-enhanced images.