1687
Residual analysis reveals variation of the intrinsic diffusivity throughout the brain in neurite orientation dispersion and density imaging (NODDI)1Medical Physics, University of Wisconsin - Madison, Madison, WI, United States, 2Waisman Center, University of Wisconsin - Madison, Madison, WI, United States, 3Psychology and Psychiatry, University of Wisconsin - Madison, Madison, WI, United States, 4Department of Computer Science, University College London, London, United Kingdom, 5Medical Physics, Psychiatry, University of Wisconsin - Madison, Madison, WI, United States
Synopsis
NOODI and its widely used estimation toolbox assume the intrinsic diffusivity to a fixed value suitable for healthy adult brains. For broader applicability of the model in neurological diseases it is important to understand the validity of assumed fixed intrinsic diffusivity. Using multi-shell diffusion data we investigated the variability of estimated NODDI indices as well as the model residuals with respect to variations in intrinsic diffusivity. The results suggest significant differences between optimum intrinsic diffusivity for white and gray matter regions as derived from intrinsic diffusivity values that generate smallest model residuals. The variability analysis indicates appreciable differences in the estimated parameters in the range of probable diffusivities predicted by the residual analysis.
Introduction
NODDI1 was developed to better characterize brain tissue microstructure from estimates of neurite volume fraction ($$$V_{ic}$$$), orientation concentration ($$$\kappa$$$), and free water (CSF) volume fraction($$$V_{iso}$$$). Since its publication in 2012, NODDI has been adopted in numerous studies for investigating microstructure in early development and other neurological conditions.2-8 One of the key fixed parameters of the NODDI model that plays a role in both optimizing acquisition protocol9 as well as in the model fitting is the intracellular parallel diffusivity ($$$d_{||}$$$) also referred as the intrinsic diffusivity. It is set to $$$1.7 μm^2ms^{-1}$$$ in the widely distributed estimation toolbox 10 as well as in protocol optimization.9 This value was estimated from the corpus callosum midsagittal plane of a healthy brain. Assuming minimal orientation dispersion, the axial diffusivity averaged over the region was estimated and used to provide an estimate for the intrinsic diffusivity. Although this assumption seems reasonable in the major tissues types of a healthy adult brain,9 as NODDI gets applied in broader settings, $$$d_{||}$$$ can vary between tissue types and pathology conditions. The main purpose of this work is to formally investigate the variability in (1) the estimated NODDI indices and (2) the accuracy of the NODDI model in terms of explaining the multi-shell diffusion data as a function of $$$d_{||}$$$.Methods
Data acquisition. Multi-shell diffusion weighted data were acquired from 5 groups of healthy volunteers (3 subjects per group, ages 1 month-72 years) on a GE 3.0 T scanner with group specific diffusion imaging parameters. Group 1 (infants): 63 directions, b: 350, 800, 1500. Group 2 (adolescents): 62 directions, b: 320, 800, 2500. Group 3 (adolescents): 57 directions, b: 500, 800, 2000. Group 4 (20-40 years): 57 directions, b: 500, 800, 2000. Group 5 (>45 years) : 105 directions, b: 300, 1200, 2700, 4800, 7500.
Analysis. NODDI indices were estimated for 16 $$$d_{||}$$$ values in the 0.5-2.0 range with a step size of 0.1 $$$μm^2ms^{-1}$$$.
Tissue segmentation. White matter (WM) and gray matter (GM) segmentations and corresponding masks where computed using FSL FAST11 on mean diffusivity and fractional anisotropy maps, which themselves where obtained by estimating the diffusion tensors from the lowest b-value diffusion weighted volumes.
Model accuracy. The relationship between model accuracy, as measured by root mean squared error (RMSE), and $$$d_{||}$$$ was examined for each voxel in all subjects. Mean RMSE values were computed across each tissue type as a function of $$$d_{||}$$$. Then, for each voxel the $$$d_{||}$$$ at which the model had the least RMSE was found, which we refer to as the optimal $$$d_{||}$$$. Optimal intrinsic diffusivity maps were segmented into WM and GM using the previously computed masks. The value of $$$d_{||}$$$ that resulted in the smallest RMSE for most voxels for each tissue type was then found by histogram analysis.
Variability of indices. For each $$$d_{||}$$$, $$$\kappa$$$, $$$V_{ic}$$$, and $$$V_{iso}$$$ maps were segmented into WM and GM. The mean for each parameter across tissue type was calculated as a function of $$$d_{||}$$$.
Results
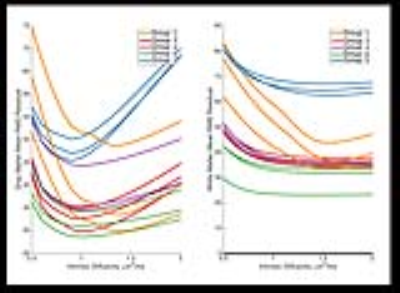
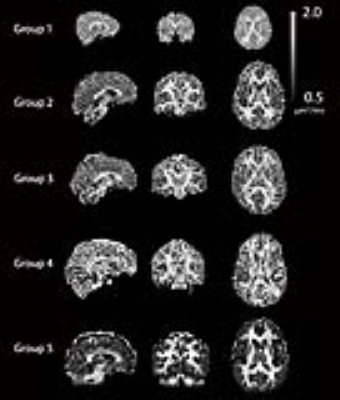
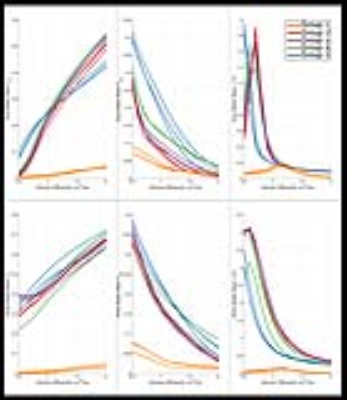
Except for the infants in Group 1, GM mean RMSE curves (Figure-1) have minima at values of $$$d_{||}$$$ that are considerably lower (<1) than the default. For Group 1, minima for the WM RMSE curves occur at values slightly less than the default. Minima in WM RMSE curves are difficult to assess for the other groups as the model residuals in WM are fairly flat for $$$d_{||}$$$ greater than 1.3. Optimal $$$d_{||}$$$ maps (Figure-2) reveal significant differences between optimum $$$d_{||}$$$ for WM and GM regions. This is confirmed with the histogram analysis (Figure-3). Compared to the default diffusivity, mode values are much lower for all groups in GM and higher in WM, except for the infants in Group 1 which also have lower than default values in WM. Figure-4 demonstrates that there are appreciable differences in the estimated parameters across the range of probable diffusivities predicted by the residual analysis [~0.7,2] for all groups.Conclusion
Our analysis of variability and model accuracy suggests that the fixed model parameter of $$$d_{||}=1.7μm^2ms^{-1}$$$ may be suboptimal and appears different for GM and WM. Further, the optimum $$$d_{||}$$$ may vary with age and protocol. This hints that, when available, a priori knowledge on $$$d_{||}$$$ should be incorporated in the estimation process by a simple two line modification of the MatLab code.7 Future work will conduct a more thorough evaluation of the effects of age and acquisition parameters on the optimal $$$d_{||}$$$ as obtained with the RMSE analysis and examine the model behavior for larger values of $$$d_{||}$$$ (up to $$$3.0 μm^2ms^{-1}$$$).Acknowledgements
This work has received support from the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1256259, the Science and Medicine Scholars Graduate Research Fellowship, and NIH grants HD090256, MH100031, and NS091733.References
- Zhang, H., Schneider, T., Wheeler-Kingshott, C.A., Alexander, D.C., 2012. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 61, 1000–1016. doi:10.1016/j.neuroimage.2012.03.072
- Billiet, T., Mädler, B., D’Arco, F., Peeters, R., Deprez, S., Plasschaert, E., Leemans, A., Zhang, H., den Bergh, B.V., Vandenbulcke, M., Legius, E., Sunaert, S., Emsell, L., 2014. Characterizing the microstructural basis of “unidentified bright objects” in neurofibromatosis type 1: A combined in vivo multicomponent T2 relaxation and multi-shell diffusion MRI analysis. NeuroImage Clin. 4, 649–658. doi:10.1016/j.nicl.2014.04.005
- Billiet, T., Vandenbulcke, M., Mädler, B., Peeters, R., Dhollander, T., Zhang, H., Deprez, S., Van den Bergh, B.R.H., Sunaert, S., Emsell, L., 2015. Age-related microstructural differences quantified using myelin water imaging and advanced diffusion MRI. Neurobiol. Aging 36, 2107–2121. doi:10.1016/j.neurobiolaging.2015.02.029
- Caverzasi, E., Papinutto, N., Castellano, A., Zhu, A.H., Scifo, P., Riva, M., Bello, L., Falini, A., Bharatha, A., Henry, R.G., 2016. Neurite Orientation Dispersion and Density Imaging Color Maps to Characterize Brain Diffusion in Neurologic Disorders: NODDI Color Map. J. Neuroimaging 26, 494–498. doi:10.1111/jon.12359
- Colgan, N., Siow, B., O’Callaghan, J.M., Harrison, I.F., Wells, J.A., Holmes, H.E., Ismail, O., Richardson, S., Alexander, D.C., Collins, E.C., Fisher, E.M., Johnson, R., Schwarz, A.J., Ahmed, Z., O’Neill, M.J., Murray,
- T.K., Zhang, H., Lythgoe, M.F., 2016. Application of neurite orientation dispersion and density imaging (NODDI) to a tau pathology model of Alzheimer’s disease. NeuroImage 125, 739–744. doi:10.1016/j.neuroimage.2015.10.043
- Kamagata, K., Hatano, T., Okuzumi, A., Motoi, Y., Abe, O., Shimoji, K., Kamiya, K., Suzuki, M., Hori, M., Kumamaru, K.K., Hattori, N., Aoki, S., 2016. Neurite orientation dispersion and density imaging in the substantia nigra in idiopathic Parkinson disease. Eur. Radiol. 26, 2567–2577. doi:10.1007/s00330-015-4066-8
- Owen, J.P., Chang, Y.S., Pojman, N.J., Bukshpun, P., Wakahiro, M.L.J., Marco, E.J., Berman, J.I., Spiro, J.E., Chung, W.K., Buckner, R.L., Roberts, T.P.L., Nagarajan, S.S., Sherr, E.H., Mukherjee, P., for the Simons VIP Consortium, 2014. Aberrant White Matter Microstructure in Children with 16p11.2 Deletions. J. Neurosci. 34, 6214–6223. doi:10.1523/JNEUROSCI.4495-13.2014
- Alexander, D.C., 2008. A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn. Reson. Med. 60, 439–448. doi:10.1002/mrm.21646
- http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab
- Zhang, Y. and Brady, M. and Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag, 20(1):45-57, 2001.
Figures