1680
Using GPUs to accelerate computational diffusion MRI: From microstructure estimation to tractography and connectomes1Oxford Centre for Functional MRI of the Brain (FMRIB), University of Oxford, Oxford, United Kingdom, 2Section for Biomedical Image Analysis (SBIA), University of Pennsylvania, Philadelphia, PA, United States, 3Oxford e-Research Centre, University of Oxford, Oxford, United Kingdom, 4Faculty of Information Technology and Bionics, Pazmany Peter Catholic University, Budapest, Hungary, 5Sir Peter Mansfield Imaging Centre, School of Medicine, University of Nottingham, Nottingham, United Kingdom
Synopsis
The great potential of computational diffusion MRI (
INTRODUCTION
The great potential of computational diffusion MRI (dMRI) relies on indirect inference of tissue microstructure and brain connections from the data. Biophysical modelling and white matter tractography frameworks map water diffusion measurements to neuroanatomical features. However, this mapping can be computationally expensive, particularly given the trend of increasing dataset sizes (for instance increased spatial, angular resolution and denser q-space sampling)1,2, the increased complexity in biophysical modelling3,4 and the large-scale imaging databases that aim to characterise variability even at the population level5,6. We have designed and developed a number of frameworks that use Graphics Processing Units (GPUs) to accelerate computations in dMRI applications. GPUs are massive parallel processors, however they are not inherently suited for all types of problems. Here, we show that, despite differences in parallelisability challenges, GPU-based designs can offer accelerations of more than two orders of magnitude, within the different contexts of tissue microstructure estimation and tractography/connectome generation.METHODS
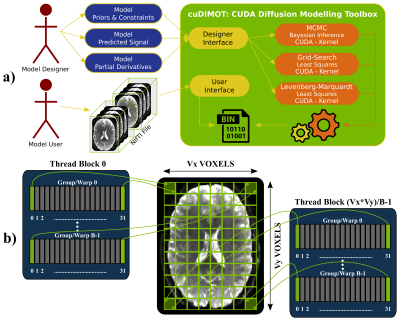
GPUs for biophysical modelling: Tissue microstructure estimation from dMRI is typically performed on a voxel-by-voxel basis, where a biophysical model is fitted7,8,9. Given the large number of voxels and the relatively low memory requirements of these independent tasks, such an application is well-suited for implementation on GPUs, which follow a SIMD (Single-Instruction over Multiple-Data streams) parallel architecture10. To take advantage of the inherent parallelisability of the problem and yet cover all possible dMRI models, we have developed a generic toolbox for designing and fitting nonlinear models using GPUs and CUDA11. The toolbox (cuDIMOT12) offers a friendly and flexible front-end for the user to implement new models (Figure-1a). The user only needs to specify a model (parameters, constraints and predicted signal) using a C language header and the translation to CUDA is then automatically performed. Various model-fitting approaches are available, including deterministic (Grid Search, nonlinear Levenberg-Marquardt optimisation), stochastic (Bayesian inference using MCMC, including a choice of likelihood functions and prior distributions) and cascaded fitting (output of a model fit is used to initialise a subsequent fitting). On top of the obvious parallelisation across voxels assigned to different threads, we found that a second level of parallelisation increases massively GPU occupancy and therefore performance. Similar to13, cuDIMOT parallelises the computation of the most expensive voxel-wise tasks, such as the evaluation of the likelihood in MCMC or the partial derivatives in Levenberg-Marquardt across different data points (Figure-1b).
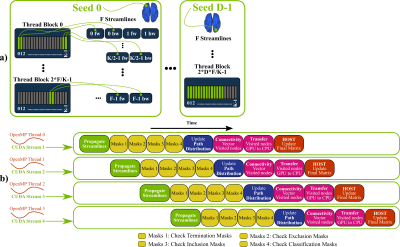
GPUs for tractography and connectome generation: Contrary to voxel-wise model fitting, white-matter tractography, and particularly whole-brain connectome generation, are not inherently suited for GPU parallelisation. Very high memory requirements, uncoalesced (non-adjacent) memory accesses and thread-divergences (irregular behaviour of threads in terms of accessed memory locations and life duration) are some of the major factors that make a GPU parallel design of such an application challenging. Nevertheless, we have developed a framework that parallelises the propagation of multiple streamlines for probabilistic tractography (Figure-2a) and overcomes the aforementioned issues using an overlapping pipelined-design (Figure-2b). This runs on a GPU by executing simultaneously multiple OpenMP threads on a CPU. Our framework can inherently handle from simple white matter tracking tasks to dense connectome generation and supports extra tractography functionality, including the ability to handle both volumes (NIFTI) and surfaces (GIFTI14), and options that allow large flexibility in imposing anatomical constraints15.
RESULTS & DISCUSSION
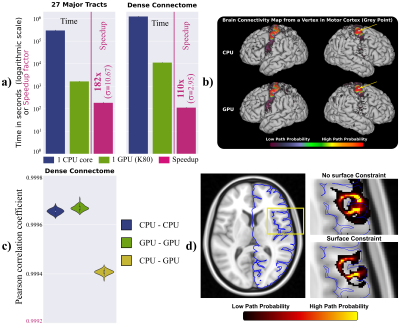
As an example for the model fitting, we used cuDIMOT to implement the NODDI3,8 model, which characterises orientation dispersion and is computationally expensive to fit to dMRI. Compared to the original NODDI Matlab toolbox8, cuDIMOT offers considerable accelerations in computation time (more than two orders of magnitude), while returning effectively identical results (Figure-3). Compared with AMICO16, a fast framework that reformulates the non-linear model fitting as a linearised system, cuDIMOT is more accurate, avoids estimation biases and is faster. The GPU tractography framework, compared with CPU implementations, achieves accelerations of over two orders of magnitude reconstructing white matter tracts or estimating a “dense” connectome (single CPU-core vs. single GPU) (Figure-4a). It also produces effectively identical results (Figure-4b-4c, correlation coefficients > 0.999). Figure-4d shows an example of the extra functionality, with the improvements obtained when using GIFTI surfaces to impose anatomical constrains in tract propagation.CONCLUSION
We have presented GPU-based frameworks for accelerating computational diffusion MRI applications, spanning from voxel-wise biophysical model fitting to whole-brain connectome generation. Despite the difference in the inherent parallelisability of these applications, GPUs can offer considerable benefits when challenges are carefully considered. Speed-ups of over two orders of magnitude change the perspective of what is computationally feasible. The GPU toolboxes will be publically released as part of FMRIB’s Software Library (FSL).Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of a Titan X Pascal GPU, which was partially used for the development of the presented toolboxes.References
- Sotiropoulos SN, Jbabdi S, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 2013; 80: 125-43.
- Setsompop K, Fan Q, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (gSlider-SMS). Magnetic Resonance in Medicine 2017.
- Tariq M, Schneider T, et al. Bingham–NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. Neuroimage 2016; 133: 207-23.
- Ferizi U, Scherrer B, et al. Diffusion MRI microstructure models with in vivo human brain Connectome data: results from a multi-group comparison. NMR in Biomedicine 2017; 30(9): 1-23.
- Glasser MF, Smith SM, et al. The Human Connectome Project’s neuroimaging approach. Nature Neuroscience 2016; 19(9): 1175–1187
- Miller KL, Alfaro-Almagro F, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience 2016; 19(11): 1523–1536
- Basser P, Mattiello J, et al. Estimation of the Effective Self-Diffusion Tensor from the NMR Spin Echo. Journal of Magnetic Resonance, Series B 1994; 103(3): 247–254.
- Zhang H, Schneider T, et al. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 2012; 61(4): 1000–16.
- Fieremans E, Jensen JH, et al. White matter characterization with diffusional kurtosis imaging. NeuroImage 2011; 58(1): 177–188.
- Flynn MJ. Some computer organizations and their effectiveness. IEEE Transaction on Computers 1972; C-21(9): 948–960.
- Nickolls J, Buck I, et al. Scalable parallel programming with CUDA. AMC Queue 2008; 6(April): 40–53.
- https://users.fmrib.ox.ac.uk/~moisesf/cudimot
- Hernández M, Guerrero G, et al. Accelerating Fibre Orientation Estimation from Diffusion Weighted Magnetic Resonance Imaging Using GPUs. PLoS ONE 2013; 8(4).
- Harwell J, Bremen H, et al. GIfTI : Geometry Data Format for Exchange of Surface-Based Brain Mapping Data. In OHBM, Melbourne, Australia 2008.
- Smith RE, Tournier JD, et al. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. NeuroImage 2012; 62(3): 1924–1938.
- Daducci A, Canales-Rodríguez EJ, et al. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. NeuroImage 2015; 105: 32–44.
Figures