1665
A Method to Quantitatively Assess and Compare Diffusion MRI Protocols between MR Systems1Radiology, University of Wisconsin, Madison, WI, United States, 2Neuroscience, University of Wisconsin, Madison, WI, United States
Synopsis
MRI systems and protocols capable of achieving diffusion measurements with comparable imaging parameters and equal or better performance to the Human Connect Project (NCP) acquisitions will enable studies in additional populations or patient groups to leverage existing HCP data as control data, decreasing costs and increasing statistical power of findings. To evaluate new MRI systems and potential protocols, we present an automated and quantitative method for evaluation of diffusion imaging performance from in-vivo data, use this method to evaluate the performance of a dMRI protocol acquired in a prototype wide bore 3T MRI system.
Introduction
Diffusion MRI (dMRI) is a powerful technique for probing microstructural integrity and structural connectivity of the brain in-vivo. The Human Connectome Project [1] (HCP) has, to date, collected and made publicly available dMRI data in 1200 healthy young adult subjects, and has additional studies underway to characterize neonatal, young adult and aging populations.
MRI systems and protocols capable of achieving diffusion measurements with comparable imaging parameters and equal or better performance to the HCP acquisitions will enable studies in additional populations or patient groups to leverage existing HCP data as control data, decreasing costs and increasing statistical power of findings. To evaluate new MRI systems and potential protocols, we present an automated and quantitative method for evaluation of diffusion imaging performance from in-vivo data, use this method to evaluate the performance of a dMRI protocol acquired in a prototype wide bore 3T MRI system, and compare the results to the HCP Lifespan 1a pilot data from the Siemens MAGNETOM Connectome Skyra 3T scanner (Siemens Healthcare, Erlangen, Germany).
Methods
Diffusion imaging was acquired in a healthy subject was scanned in accordance to local ethics guidelines on a prototype wide bore GE Signa Premier 3T (GE Healthcare, Waukesha, WI, USA) with gradient strength of 80 mT/m and peak slew rate of 200 T/m/s. Relevant imaging parameters are 1.5mm isotropic resolution, simultaneous multislice (HyperBand) acceleration factor HB3, no parallel imaging, b=1500,3000 s/mm2, 92 directions per shell, 16 b=0 images, and repeated acquisition with AP and PA phase encoding.
Comparison data used in the preparation of this work were obtained from the Human Connectome Project (HCP) database (https://ida.loni.usc.edu/login.jsp). A single subject matched in sex and age range was selected from the Lifespan 1a pilot data release, acquired on the Connectome Skyra 3T scanner with b=1000,2500s/mm2, 75 directions per shell, 10 b=0 images, LR and RL phase encoding, and other parameters identical to those listed above.
Data were preprocessed using a simplified version of the HCP diffusion pipeline, including distortion correction with FSL TOPUP [2], correction for motion and eddy currents with FSL Eddy [3], and diffusion tensor fitting. Estimation of crossing fibers was performed using BEDPOSTX [4].
DTI fractional anisotropy (FA) images were nonlinearly registered using FNIRTFSL to the FMRIB58 diffusion space to define white matter regions of interest (ROIs). Temporal and fiber orientation uncertainty and fraction of voxels with 2-way and 3-way fiber crossings were computed [5] from the BEDPOSTX estimates to objectively assess data quality.
Results and Discussion
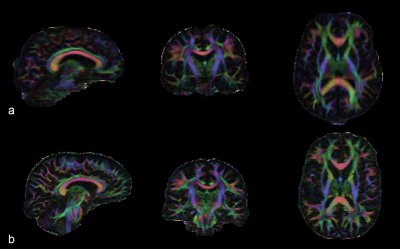
dMRI data collected on the wide-bore 3T system was compatible with the standard HCP preprocessing pipeline, and the quality of distortion, motion, and eddy current corrections were visually comparable to existing HCP pilot data (Figure 1). The protocol implemented on the test system, with a maximum b-value of 3000s/mm2, achieved an echo time of 69ms and echo spacing of 736µs, while the Connectome system with the same b-value is able to achieve 76ms and 690µs, respectively. Short echo time is critically important in high b-value imaging to prevent loss of signal due to T2 decay, while echo spacing is important to minimize distortions, especially when in-plane parallel imaging is not used.
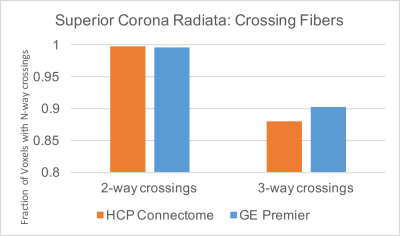
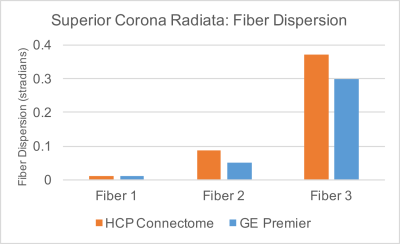
Quantitative evaluation of the two systems and protocols in the superior corona radiata, a white matter region with many crossing fiber pathways, revealed 99.7% of voxels support 2-way and 90.2% support 3-way crossings on the wide-bore 3T protocol, compared to 99.8% and 88.0%, respectively, for the HCP Lifespan 1a protocol (Figure 2). Angular uncertainty in fiber estimates (lower is better) were 0.010, 0.051, and 0.298 stradians for the first, second, and third fiber population, respectively, for the wide-bore protocol and 0.013, 0.086, and 0.37 stradians for the HCP Lifespan 1a protocol (Figure 3). Based on these metrics, the wide-bore 3T system is capable of meeting the performance necessary for HCP-compatible acquisitions.
Conclusions
We present an objective method to compare the quality of diffusion MRI data across different protocols and systems, by evaluating the quality of crossing fiber estimation in atlas-defined white matter regions. We demonstrate the method by comparing the performance of two different protocols across two different systems in a single volunteer. By including a larger number of neurotypical volunteers, this approach can be used to compare different MRI systems, and diffusion protocols within system. The example presented is limited by the fact that only one (different) subject was scanned, and that the protocol on the test system contained more sampled directions than the HCP protocol. Future work will test protocols matched in both number of samples and scan time to better establish equivalence with HCP datasets.Acknowledgements
No acknowledgement found.References
[1] David C. Van Essen, Stephen M. Smith, Deanna M. Barch, Timothy E.J. Behrens, Essa Yacoub, Kamil Ugurbil, for the WU-Minn HCP Consortium. (2013). The WU-Minn Human Connectome Project: An overview. NeuroImage 80(2013):62-79
[2] S.M. Smith, M. Jenkinson, M.W. Woolrich, C.F. Beckmann, T.E.J. Behrens, H. Johansen-Berg, P.R. Bannister, M. De Luca, I. Drobnjak, D.E. Flitney, R. Niazy, J. Saunders, J. Vickers, Y. Zhang, N. De Stefano, J.M. Brady, and P.M. Matthews. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23(S1):208-219, 2004.
[3] Jesper L. R. Andersson and Stamatios N. Sotiropoulos. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage, 125:1063-1078, 2016.
[4] T.E.J. Behrens, M.W. Woolrich, M. Jenkinson, H. Johansen-Berg, R.G. Nunes, S. Clare, P.M. Matthews, J.M. Brady, and S.M. Smith. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med, 50(5):1077-1088, 2003.
[5] Lifespan Pilot Report, 10 Feb 2016 (https://www.humanconnectome.org/storage/app/media/documentation/lifespan-pilot/WU-Minn_HCP_LifespanPilot_Report_10Feb2015.pdf)
Figures