1663
A comparison of different brain connectivity markers for classifying Gulf-war illness1Anatomy and Neurobiology, Boston University, Boston, MA, United States, 2Boston University School of Public Health, Boston, MA, United States
Synopsis
Gulf War Illness (GWI) represents a cluster of multi-system chronic symptoms experienced by a third of veterans who served in the Gulf War. The exact cause of GWI remains unknown and efforts directed towards developing treatments have been hampered by the lack of meaningful objective biomarkers of the illness. Combining machine learning technology to brain connectivity imaging may allow for better understanding of the complex pathobiology of GWI. Choosing optimal imaging index should be a first step to maximize its classification performance.
Introduction
Symptoms of Gulf War illness (GWI) typically include persistent headaches, widespread pain, fatigue, memory and concentration problems and other difficulties in this complex multi-system disorder. Converging evidence suggests that a clear understanding of brain-immune interactions can help us to understand the origin of these symptoms. Morphometric neuroimaging analysis on Gulf War (GW) veterans exposed to neurotoxic and nerve agents confirmed overall reduction in the grey matter (GM)1 and white matter (WM)2,3, compared to the non exposed veterans. These regional changes in the morphometry of various brain regions have also been tied to changes in brain connectivity using diffusion MRI4,5. These findings may indicate that there are focal spots primarily involved in the illness propagation in the brain. In this study, we have applied a machine learning framework to diffusion magnetic resonance imaging markers from gulf war veterans to assess brain connections specific to Gulf-War illness (GWI).Methods
Training set was based on 20 GW veterans (8 GW controls and 12 GWI, based on Kansas criteria) and 18 GW veterans’ data was used for the test set. We also included non-veteran aged control data (12 controls) in this study which have the same diffusion MRI data. Brain structural network was extracted from high angular resolution diffusion imaging (HARDI, spin-echo epi, 1.75/1.75/2mm voxel, 65 independent diffusion gradient directions, b-value 3000s/mm2) data collected in the Boston Gulf War Illness Consortium (GWIC). Local Brain connection was defined based on the existence white matter tracts between each of the GM regions of interests (Freesufer ROIs, total 78). Per each local brain connection, we applied the following quantifications for defining local connectivity measures: 1) total number of tracts, 2) tract mean value of microscale diffusivity6 from generalized q-space reconstruction. We applied random decision forest classification for machine learning of the brain connectivity. Machine learning classifier was trained for different quantifications and compared with each other. Performance of the machine learning classifier was tested based on a leave one out cross validation (LOOCV) and test set classification performance.Results
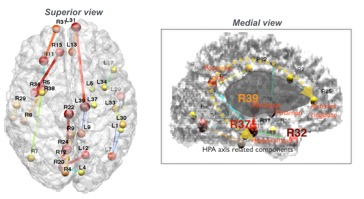
The highest classification performance in GW controls/GWI was confirmed in microscale diffusivity quantification. Classification based on the microscale diffusivity revealed an accuracy of 84% in the LOOCV. In the test data validation, it showed 77% accuracy of classifying GWI. From the microscale diffusivity based classification, veterans with GWI had significantly different connectional alterations in several cortical regions compared to control veterans (Figure). Important connection features extracted by the classification model were found in the regions including ‘the thalamus (L39) - frontal pole (L31)’, ‘the posterior cingulate (R22) - precuneous (R24)’, ‘the hippocampus (L37) - thalamus (L39)’ and ‘the insula (R34) - medial orbitofrontal (R13)’ connections. Classification based on the total number of tracts revealed an accuracy of 54% in the LOOCV. In the test data validation, it showed 46% accuracy of classifying GWI. Classifying between GWI and non-veteran aged control based on the total number of connections showed over 90% in both LOOCV and test dataset classification.Discussions
Brain connectomic techniques have the potential power to uncover the underlying mechanisms of GWI. The advantage of the brain connectomics lies in their capacity to map effects of interest in both focal as well as a large-scale data points. Brain connectomics allows for processing of a large and distributed number of brain connections as well as more local and focal connections. However, our preliminary data showed that different quantification strategies in diffusion MRI can have significant impact on describing group characteristics. Lower range of diffusion encodings used in diffusion MRI (typically, around b=1000s/mm2) is the most common set up in the clinical imaging and draws mostly the fast diffusion components and it might not have enough sensitivity to detect microscopic water diffusivity changes. Better sensitivity for detecting micro-diffusivity can be obtained by adding high diffusion strength encoding (typically, b>1,800s/mm2). As shown in our preliminary observations, micro-diffusion can be a sensitive index to more diffuse changes in the brain. Since latent risk factors can possibly be present in some of the GW controls, it is understandable that classifying GWI from the non-veteran control samples could have better performances compared to the other one. However, more data is needed for concrete conclusion to be made.Conclusion
Combining machine learning technology to brain connectivity imaging may allow for better understanding of the complex pathobiology of GWI. Choosing optimal imaging index should be a first step to maximize its classification performance. We are now extending our work by adding blood cytokine and cognitive measurements to the existing neuroimaging data to test multimodal data classification of GWI.Acknowledgements
This work is supported by a department of Defense CDMRP GWI consortium award (W81XWH-17-1-0440). Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.References
[1] Chao LL, Rothlind JC, Cardenas VA, Meyerhoff DJ, Weiner MW.Effects of low-level exposure to sarin and cyclosarin during the 1991 Gulf War on brain function and brain structure in US veterans. Neurotoxicology 2010;31:493–501.
[2] Heaton KJ, Palumbo CL, Proctor SP, Killiany RJ, Yurgelun-Todd DA, White RF. Quantita- tive magnetic resonance brain imaging in US army veterans of the 1991 Gulf War potentially exposed to sarin and cyclosarin. Neurotoxicology 2007;28:761–9.
[3] Chao LL, Abadjian L, Hlavin J, Meyerhoff DJ, Weiner MW.Effects of low-level sarin and cyclosarin exposure and Gulf War Illness on brain structure and function: a study at 4 Tesla. Neurotoxicology 2011;32:814–22.
[4] Rayhan, R. U., Stevens, B. W., Timbol, C. R., Adewuyi, O., Walitt, B., VanMeter, J. W., et al. (2013). Increased brain white matter axial diffusivity associated with fatigue, pain and hyperalgesia in Gulf War illness [Research Support, N.I.H., Extramural, U.S. Gov't, Non-P.H.S.] PLoS One, 8(3), e58493.
[5] Chao, L. L., Zhang, Y., & Buckley, S. (2015). Effects of low-level sarin and cyclosarin exposure on white matter integrity in Gulf War Veterans. NeuroToxicology, 48, 239–248.
[6] Yeh, Fang-Cheng, Li Liu, T. Kevin Hitchens, and Yijen L. Wu, "Mapping Immune Cell Infiltration Using Restricted Diffusion MRI", Magn Reson Med. accepted, (2016)