1637
Effects of phase error on image reconstruction for simultaneous multi-slice readout-segmented diffusion MRI1ECE, University of Utah, SALT LAKE CITY, UT, United States, 2Radiology and Imaging Sciences, University of Utah, SALT LAKE CITY, UT, United States, 3Bioengineering, University of Utah, SALT LAKE CITY, UT, United States, 4Biomedical Engineering, The State University of New York (SUNY) at Buffalo, Buffalo, NY, United States
Synopsis
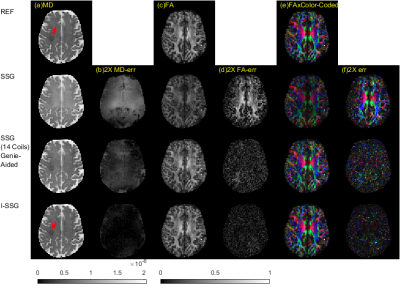
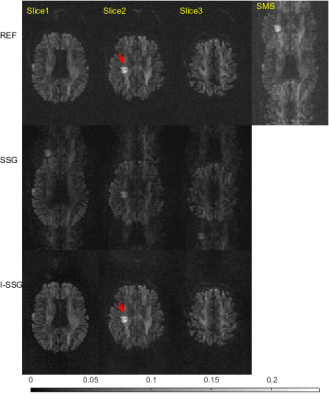
In this work, we study the effect of phase errors on the quality of image reconstructions for simultaneous multi-slice (SMS) readout-segmented echo planar imaging (RS-EPI) acquisitions. We propose an iterative split slice-GRAPPA (I-SSG) algorithm to train improved kernels using estimated diffusion weighted images (DWIs) rather than baseline images. Results from stroke patients show that the proposed I-SSG algorithm produces consistently better reconstructions than the SSG algorithm in the presence of baseline phase errors.
Introduction
Readout-segmented echo planar imaging (RS-EPI) [1] combined with simultaneous multi-slice (SMS) acquisition with blipped-controlled aliasing in parallel imaging (CAIPI) [2,3] improves spatial resolution of diffusion-weighted images (DWIs) and reduces scan time for both single-shot [2] and RS-EPI [4]. Slice-GRAPPA [2] and its improved version split slice-GRAPPA (SSG) [5] are commonly used methods to de-alias SMS DWIs using kernels trained from baseline images. When applying SSG to datasets acquired from a RS-EPI sequence, we found that SSG kernels trained from baselines do not de-alias DWIs effectively due to baseline phase errors.
To overcome this issue, we propose an iterative approach, termed iterative split slice-GRAPPA (I-SSG), to train improved kernels using estimated DWIs rather than baseline images. Our results from a stroke patient show that proposed I-SSG produces consistently better reconstructions in the presence of baseline phase errors. It yields over 50% improvement over SSG in fractional anisotropy (FA) and mean diffusion (MD) estimations for slice reduction factors of up to R=4.
Theory and methods
We consider multi-coil and multi-slice DWIs with $$$N_c$$$ coils and $$$N_s$$$ simultaneous slices. The DWIs for the $$$i$$$-th coil, $$$l$$$-th slice, and $$$n$$$-th diffusion direction are written as
$$m_{i,l,n}(\vec{x})=|s_{l}(\vec{x})||c_{i,l}(\vec{x})|e^{j\theta_{i,l,n}(\vec{x})}a_{l,n}(\vec{x}) \quad (1)$$
where $$$\vec{x}$$$ is pixel position, $$$|s_{l}|$$$ is magnitude of baseline, $$$|c_{i,l}|$$$ is magnitude of coil sensitivity, $$$\theta_{i,l,n}$$$ is the phase, and $$$a_{l,n}$$$ is the attenuation due to diffusion.
Assume that $$$N_s$$$ phase modulated slices are acquired simultaneously according to CAIPI scheme [3] . Let superscript $$$(\phi_l)$$$ denote phase modulation of $$$l$$$-th slice. The SMS image is summation of phase modulated images, written as $$\label{eq-sms}r_{i,n} =\textstyle\sum_{l=1}^{N_s}m_{i,l,n}^{(\phi_l)}=\textstyle\sum_{l=1}^{N_s}(|s_{l}||c_{i,l}|e^{j\theta_{i,l,n}})^{(\phi_l)}a_{l,n}^{(\phi_l)} \quad (2).$$
In SSG, k-space samples of single slices are estimated by applying kernels on SMS data. The key idea of I-SSG is to use estimated DWIs, rather than baselines, to re-train kernels over time. Main steps of I-SSG are as follows. Step1. Initialization. Non-SMS baseline images are used to train initial SSG kernels. Step 2. K-space de-aliasing. Current kernels are used to de-alias SMS data in k-space and estimate single slice images $$$ \widehat{m}_{i,l,n}$$$. Step 3. Image domain de-aliasing. We estimate $$$|s_{l}||c_{i,l}|$$$ in (1) by magnitude of baseline multi-coil images without estimating coil sensitivities. Furthermore, we constrain the phase of each DWI to be the phase of SSG output from Step 2, i.e., $$$\widehat{\theta}_{i,l,n}=\measuredangle\widehat{m}_{i,l,n}$$$. From (2), we obtain the following linear system of equations.
$$\begin{bmatrix}r_{1,n} \\r_{2,n} \\\vdots \\r_{Nc,n}\end{bmatrix} =\begin{bmatrix}(|s_{1}||c_{1,1}|e^{j\widehat{\theta}_{1,1,n}})^{(\phi_l)} & \dots & (|s_{N_s}||c_{1,N_s}|e^{j\widehat{\theta}_{1,N_s,n}})^{(\phi_{N_s})}\\(|s_{1}||c_{2,1}|e^{j\widehat{\theta}_{2,1,n}})^{(\phi_1)} & \dots & (|s_{N_s}||c_{2,N_s}|e^{j\widehat{\theta}_{2,N_s,n}})^{(\phi_{N_s})} \\\vdots & \ddots & \vdots \\(|s_{1}||c_{N_c,1}|e^{j\widehat{\theta}_{N_c,1,n}})^{(\phi_1)} & \dots & (|s_{N_s}||c_{N_c,N_s}|e^{j\widehat{\theta}_{N_c,N_s,n}})^{(\phi_{N_s})}\end{bmatrix}\begin{bmatrix}\widehat{a}_{1,n}^{(\phi_1)} \\\widehat{a}_{2,n}^{(\phi_2)} \\\vdots \\\widehat{a}_{N_s,n}^{(\phi_{N_s})}\end{bmatrix} \quad (3)$$
Writing (3) in a matrix form $$$\mathbf{r}_n=\mathbf{A}_n\widehat{\mathbf{a}}_n^{(\phi)}$$$, we have
$$\label{eq-atten-est}\widehat{\mathbf{a}}_n^{(\phi)}=(\mathbf{A}_n^H\mathbf{A}_n)^{-1}\mathbf{A}_n^H\mathbf{r}_n. \quad (4)$$
Having estimated all components in (1), we update DWIs as
$$\label{dwi-est-1}\breve{m}_{i,l,n} = |s_{l}||c_{i,l}|e^{j\widehat{\theta}_{i,l,n}}\mathscr{F}^{-1}\{e^{-j\phi_l}\mathscr{F}\{\widehat{a}_{l,n}^{(\phi_l)}\}\} \quad (5)$$ where $$$\mathscr{F}\{.\}$$$ denotes 2D discrete Fourier transform.
Step 4. Tensor fitting. We combine $$$\breve{m}_{i,l,n}$$$ from (5) using sum of squares to obtain $$$\widetilde{m}_{l,n}=\sqrt{\textstyle\sum_{i=1}^{N_c}|\breve{m}_{i,l,n}|^2}$$$. The diffusion matrices are then estimated using $$\label{tensor-fitting} \widehat{D}_l,\widehat{|s_l|}=\underset{D_l,|s_l|}{\arg\min}\textstyle\sum_n||\widetilde{m}_{l,n}-|s_{l}|e^{-bg_n^TD_lg_n}||^2\text{ s.t. }D_l\succ 0, \quad (6)$$ where $$$D_l$$$ is diffusion tensor matrix, $$$g_n$$$ is $$$n$$$-th diffusion direction. Step 5. Re-train kernels. Estimates of DWIs are refined by $$\label{dwi-est-2} \check{m}_{i,l,n} = |s_{l}||c_{i,l}|e^{j\measuredangle\breve{m}_{i,l,n}}e^{-bg_n^T\widehat{D}_lg_n}. \quad (7)$$
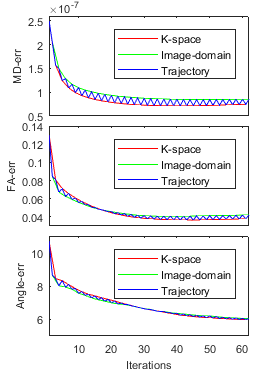
We re-train new kernels for SSG using DWIs from (7) corresponding to diffusion direction $$$n=n_0 $$$ that has minimum mean-square-error against acquired SMS data. The tensor fitting step improves kernels by effectively de-noising estimated DWIs.
Step 6. Repeat. Go to Step 2 until algorithm converges.
Results
We used fully sampled RS-EPI data acquired from a Siemens 3T Verio scanner with 32-channel head coils. A stroke patient dataset with IRB approval and informed consent was processed. One b=0 and twenty DWIs with a b-value of 2000 s/mm2 were acquired with TR=3.5 secs, TE=100 msec, 7 shots, 16 slices with slice thickness=2.1 mm, and pixel size=2.1 x 2.1 mm2. Nyquist corrections, regridding, and navigator corrections were done according to [1]. SMS RS-EPI data with slice acceleration factor R=2,3,4 were simulated from pre-processed RS-EPI data by phase modulating the slices and adding them together.
Superior performance of proposed I-SSG over SSG are shown in Table1, Table2, Fig. 1, and Fig. 2. Fig. 3 shows the convergence of I-SSG over iterations. Similar results (not shown here) were obtained for two other datasets of stroke patients.
Conclusions
To the best of our knowledge, this is the first work to investigate the impact of phase mismatch between training data and actual DWI data on the effectiveness of SSG for SMS reconstructions. Our results demonstrated that I-SSG significantly reduces FA, MD, and angle errors compared to SSG in the presence of phase errors. The approach of iteratively re-training SSG kernels using estimated DWIs is unique and might find further applications in other SMS acquisitions where phase inconsistencies exist.Acknowledgements
This work was supported by University of Utah Seed Grant, NIHgrants R01NS083761, R21EB020861, and R01EB025133-01. We thank Dr. Steen Moller for providing us with codes for SSG and Dr.Robert Frost for the codes to read in RESOLVE data.References
[1] D. A. Porter and R. M. Heidemann, “High resolution diffusion-weightedimaging using readout-segmented echo-planar imaging, parallel imagingand a two-dimensional navigator-based reacquisition,” Magnetic Resonancein Medicine, vol. 62, no. 2, pp. 468–475, 2009.
[2] K. Setsompop, B. A. Gagoski, J. R. Polimeni, T. Witzel, V. J. Wedeen,and L. L. Wald, “Blipped-controlled aliasing in parallel imaging for simultaneousmultislice echo planar imaging with reduced g-factor penalty,”Magnetic Resonance in Medicine, vol. 67, no. 5, pp. 1210–1224,2012.
[3] F. A. Breuer, M. Blaimer, R. M. Heidemann, M. F. Mueller, M. A. Griswold,and P. M. Jakob, “Controlled aliasing in parallel imaging resultsin higher acceleration (caipirinha) for multi-slice imaging,” MagneticResonance in Medicine, vol. 53, no. 3, pp. 684–691, 2005.
[4] R. Frost, P. Jezzard, G. Douaud, S. Clare, D. A. Porter, and K. L. Miller,“Scan time reduction for readout-segmented epi using simultaneous multisliceacceleration: Diffusion-weighted imaging at 3 and 7 tesla,” MagneticResonance in Medicine, vol. 74, no. 1, pp. 136–149, 2015.
[5] S. F. Cauley, J. R. Polimeni, H. Bhat, L. L. Wald, and K. Setsompop,“Interslice leakage artifact reduction technique for simultaneous multisliceacquisitions,” Magnetic Resonance in Medicine, vol. 72, no. 1, pp.93–102, 2014.
Figures